
Capybara microvirus Cap1_SP_61
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
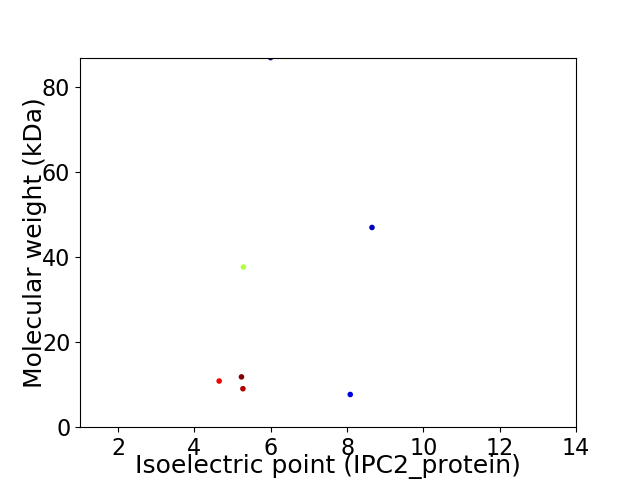
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
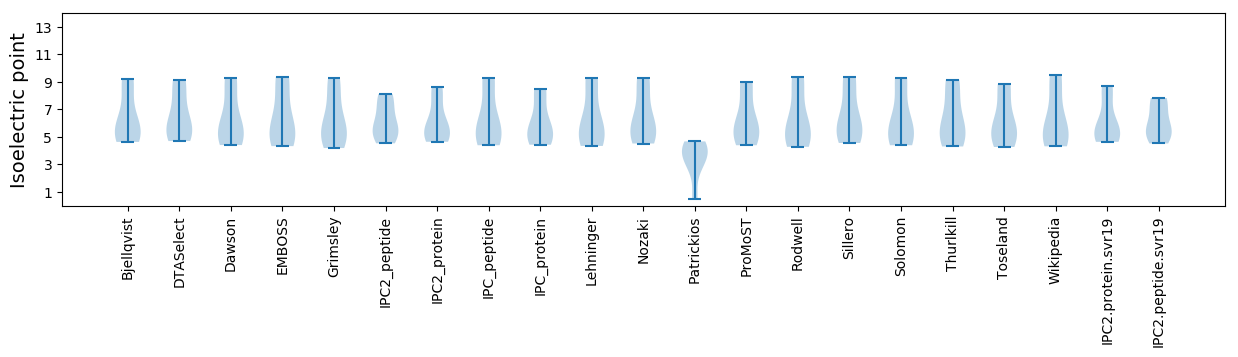
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W3X3|A0A4P8W3X3_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_61 OX=2584786 PE=4 SV=1
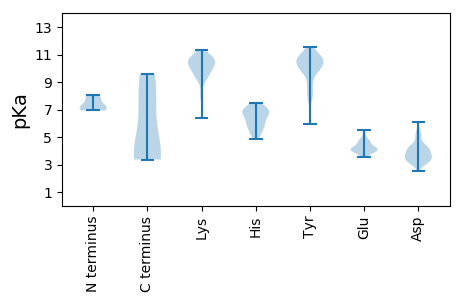
MM1 pKa = 7.93RR2 pKa = 11.84FTNNFVFAHH11 pKa = 5.57PVFLNSNNTPTAQGASLTPAQIAKK35 pKa = 9.06ATMEE39 pKa = 5.21GIPISPQGLSTEE51 pKa = 4.46QYY53 pKa = 10.75FEE55 pKa = 4.92GNTGDD60 pKa = 3.45SMYY63 pKa = 10.34IAPQFMRR70 pKa = 11.84GVDD73 pKa = 5.52LNDD76 pKa = 2.93AWNIVQDD83 pKa = 3.82NKK85 pKa = 11.18KK86 pKa = 10.32SFNSLTISEE95 pKa = 4.77NGNDD99 pKa = 3.36
MM1 pKa = 7.93RR2 pKa = 11.84FTNNFVFAHH11 pKa = 5.57PVFLNSNNTPTAQGASLTPAQIAKK35 pKa = 9.06ATMEE39 pKa = 5.21GIPISPQGLSTEE51 pKa = 4.46QYY53 pKa = 10.75FEE55 pKa = 4.92GNTGDD60 pKa = 3.45SMYY63 pKa = 10.34IAPQFMRR70 pKa = 11.84GVDD73 pKa = 5.52LNDD76 pKa = 2.93AWNIVQDD83 pKa = 3.82NKK85 pKa = 11.18KK86 pKa = 10.32SFNSLTISEE95 pKa = 4.77NGNDD99 pKa = 3.36
Molecular weight: 10.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVM8|A0A4V1FVM8_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_61 OX=2584786 PE=4 SV=1
MM1 pKa = 7.47ICLSPKK7 pKa = 9.58YY8 pKa = 9.8SLNYY12 pKa = 9.79RR13 pKa = 11.84ILGKK17 pKa = 10.3IMPNIVGYY25 pKa = 10.21SIEE28 pKa = 4.07NQVYY32 pKa = 7.41FTKK35 pKa = 10.39PSYY38 pKa = 10.58RR39 pKa = 11.84SLQCALDD46 pKa = 3.77DD47 pKa = 5.11LIAFGSSSYY56 pKa = 10.7KK57 pKa = 10.62VLVTGYY63 pKa = 10.32YY64 pKa = 10.52DD65 pKa = 3.49GLSVVNYY72 pKa = 8.22PVSPFIVTPCGHH84 pKa = 6.85CASCLKK90 pKa = 10.64AKK92 pKa = 9.96QLNKK96 pKa = 6.35TTCMQLEE103 pKa = 4.3ALNYY107 pKa = 7.95RR108 pKa = 11.84QPLFVTLTYY117 pKa = 10.85ADD119 pKa = 4.45EE120 pKa = 4.82PPSGVNVKK128 pKa = 10.16DD129 pKa = 3.35VQNFLKK135 pKa = 10.64RR136 pKa = 11.84LRR138 pKa = 11.84TNLKK142 pKa = 9.73RR143 pKa = 11.84KK144 pKa = 10.23GYY146 pKa = 9.64DD147 pKa = 2.98EE148 pKa = 5.53PIRR151 pKa = 11.84YY152 pKa = 8.78HH153 pKa = 5.94CTAEE157 pKa = 4.25YY158 pKa = 10.69GSKK161 pKa = 9.29TYY163 pKa = 10.2RR164 pKa = 11.84PHH166 pKa = 5.22YY167 pKa = 10.08HH168 pKa = 6.95LIIYY172 pKa = 8.21NMPNEE177 pKa = 4.25EE178 pKa = 4.47FFDD181 pKa = 4.1LPAFGSYY188 pKa = 9.96GAGYY192 pKa = 10.35AYY194 pKa = 10.51DD195 pKa = 4.53DD196 pKa = 5.51FIRR199 pKa = 11.84SCWKK203 pKa = 10.52LGLTHH208 pKa = 6.66SRR210 pKa = 11.84LVSDD214 pKa = 4.15VAALRR219 pKa = 11.84YY220 pKa = 8.0CAKK223 pKa = 9.35YY224 pKa = 7.8TAKK227 pKa = 10.36CSSILKK233 pKa = 9.89GKK235 pKa = 9.8NPPFMLQSTKK245 pKa = 10.98YY246 pKa = 9.45GGLGSKK252 pKa = 9.77VYY254 pKa = 10.79KK255 pKa = 10.68NLANEE260 pKa = 4.53FLTQKK265 pKa = 10.88NIVNVNGKK273 pKa = 8.14QVSLPLNAYY282 pKa = 7.86FLRR285 pKa = 11.84KK286 pKa = 9.47VLPSAQQCVGYY297 pKa = 9.86YY298 pKa = 8.05WLRR301 pKa = 11.84IMKK304 pKa = 10.16SLEE307 pKa = 4.07LCGKK311 pKa = 9.86LPPLHH316 pKa = 6.83PFYY319 pKa = 10.72IDD321 pKa = 4.18LLHH324 pKa = 7.0HH325 pKa = 5.74YY326 pKa = 10.15QPAKK330 pKa = 10.74DD331 pKa = 3.48SAIYY335 pKa = 10.55ADD337 pKa = 4.03IVILSGQILKK347 pKa = 8.2TCYY350 pKa = 9.79YY351 pKa = 9.83RR352 pKa = 11.84GLISSRR358 pKa = 11.84FTDD361 pKa = 5.14FSAQKK366 pKa = 9.62TIEE369 pKa = 3.97FLNFKK374 pKa = 10.53HH375 pKa = 6.99SFLQNLPYY383 pKa = 10.29QSPAEE388 pKa = 3.96PKK390 pKa = 10.06ILEE393 pKa = 4.11NYY395 pKa = 8.57IAQNQLDD402 pKa = 3.89RR403 pKa = 11.84QKK405 pKa = 10.82EE406 pKa = 4.11KK407 pKa = 11.15EE408 pKa = 3.99IMM410 pKa = 3.8
MM1 pKa = 7.47ICLSPKK7 pKa = 9.58YY8 pKa = 9.8SLNYY12 pKa = 9.79RR13 pKa = 11.84ILGKK17 pKa = 10.3IMPNIVGYY25 pKa = 10.21SIEE28 pKa = 4.07NQVYY32 pKa = 7.41FTKK35 pKa = 10.39PSYY38 pKa = 10.58RR39 pKa = 11.84SLQCALDD46 pKa = 3.77DD47 pKa = 5.11LIAFGSSSYY56 pKa = 10.7KK57 pKa = 10.62VLVTGYY63 pKa = 10.32YY64 pKa = 10.52DD65 pKa = 3.49GLSVVNYY72 pKa = 8.22PVSPFIVTPCGHH84 pKa = 6.85CASCLKK90 pKa = 10.64AKK92 pKa = 9.96QLNKK96 pKa = 6.35TTCMQLEE103 pKa = 4.3ALNYY107 pKa = 7.95RR108 pKa = 11.84QPLFVTLTYY117 pKa = 10.85ADD119 pKa = 4.45EE120 pKa = 4.82PPSGVNVKK128 pKa = 10.16DD129 pKa = 3.35VQNFLKK135 pKa = 10.64RR136 pKa = 11.84LRR138 pKa = 11.84TNLKK142 pKa = 9.73RR143 pKa = 11.84KK144 pKa = 10.23GYY146 pKa = 9.64DD147 pKa = 2.98EE148 pKa = 5.53PIRR151 pKa = 11.84YY152 pKa = 8.78HH153 pKa = 5.94CTAEE157 pKa = 4.25YY158 pKa = 10.69GSKK161 pKa = 9.29TYY163 pKa = 10.2RR164 pKa = 11.84PHH166 pKa = 5.22YY167 pKa = 10.08HH168 pKa = 6.95LIIYY172 pKa = 8.21NMPNEE177 pKa = 4.25EE178 pKa = 4.47FFDD181 pKa = 4.1LPAFGSYY188 pKa = 9.96GAGYY192 pKa = 10.35AYY194 pKa = 10.51DD195 pKa = 4.53DD196 pKa = 5.51FIRR199 pKa = 11.84SCWKK203 pKa = 10.52LGLTHH208 pKa = 6.66SRR210 pKa = 11.84LVSDD214 pKa = 4.15VAALRR219 pKa = 11.84YY220 pKa = 8.0CAKK223 pKa = 9.35YY224 pKa = 7.8TAKK227 pKa = 10.36CSSILKK233 pKa = 9.89GKK235 pKa = 9.8NPPFMLQSTKK245 pKa = 10.98YY246 pKa = 9.45GGLGSKK252 pKa = 9.77VYY254 pKa = 10.79KK255 pKa = 10.68NLANEE260 pKa = 4.53FLTQKK265 pKa = 10.88NIVNVNGKK273 pKa = 8.14QVSLPLNAYY282 pKa = 7.86FLRR285 pKa = 11.84KK286 pKa = 9.47VLPSAQQCVGYY297 pKa = 9.86YY298 pKa = 8.05WLRR301 pKa = 11.84IMKK304 pKa = 10.16SLEE307 pKa = 4.07LCGKK311 pKa = 9.86LPPLHH316 pKa = 6.83PFYY319 pKa = 10.72IDD321 pKa = 4.18LLHH324 pKa = 7.0HH325 pKa = 5.74YY326 pKa = 10.15QPAKK330 pKa = 10.74DD331 pKa = 3.48SAIYY335 pKa = 10.55ADD337 pKa = 4.03IVILSGQILKK347 pKa = 8.2TCYY350 pKa = 9.79YY351 pKa = 9.83RR352 pKa = 11.84GLISSRR358 pKa = 11.84FTDD361 pKa = 5.14FSAQKK366 pKa = 9.62TIEE369 pKa = 3.97FLNFKK374 pKa = 10.53HH375 pKa = 6.99SFLQNLPYY383 pKa = 10.29QSPAEE388 pKa = 3.96PKK390 pKa = 10.06ILEE393 pKa = 4.11NYY395 pKa = 8.57IAQNQLDD402 pKa = 3.89RR403 pKa = 11.84QKK405 pKa = 10.82EE406 pKa = 4.11KK407 pKa = 11.15EE408 pKa = 3.99IMM410 pKa = 3.8
Molecular weight: 46.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1873 |
68 |
774 |
267.6 |
30.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.46 ± 0.832 | 1.548 ± 0.506 |
4.645 ± 0.379 | 4.698 ± 1.014 |
4.645 ± 0.774 | 5.392 ± 0.353 |
1.762 ± 0.408 | 6.941 ± 0.52 |
5.072 ± 0.834 | 8.916 ± 0.943 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.403 ± 0.255 | 7.208 ± 0.702 |
4.592 ± 0.69 | 4.592 ± 0.494 |
4.164 ± 0.295 | 8.97 ± 0.488 |
6.407 ± 0.661 | 5.392 ± 0.676 |
0.481 ± 0.17 | 5.713 ± 0.972 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
