
Methylobacterium sp. TER-1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Methylobacterium; unclassified Methylobacterium
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
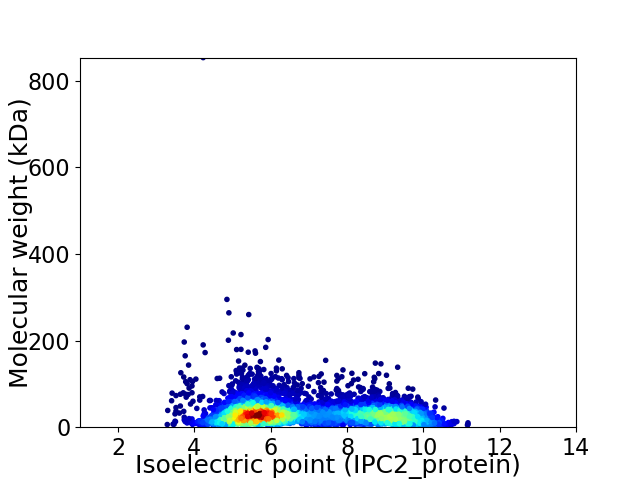
Virtual 2D-PAGE plot for 5625 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S2VK25|A0A3S2VK25_9RHIZ EAL domain-containing protein OS=Methylobacterium sp. TER-1 OX=2499852 GN=EOE48_22345 PE=4 SV=1
MM1 pKa = 7.5ATTSYY6 pKa = 11.18YY7 pKa = 10.49FGYY10 pKa = 10.64LNRR13 pKa = 11.84NEE15 pKa = 4.37PLNTYY20 pKa = 9.27TYY22 pKa = 10.44LSQYY26 pKa = 10.83ADD28 pKa = 3.9TITALNNGGFFTAYY42 pKa = 9.47EE43 pKa = 4.57GYY45 pKa = 10.82YY46 pKa = 10.6DD47 pKa = 4.1GGYY50 pKa = 10.79NYY52 pKa = 10.19ILARR56 pKa = 11.84GLDD59 pKa = 3.75ATGAPAPKK67 pKa = 10.64GEE69 pKa = 4.26FAATNYY75 pKa = 10.49GPGINVRR82 pKa = 11.84SAEE85 pKa = 3.95AATLKK90 pKa = 10.7NGNVVMTYY98 pKa = 7.04TTQSSTSTPITFRR111 pKa = 11.84IFSPDD116 pKa = 2.91GTLLTEE122 pKa = 4.96GSTASGSNYY131 pKa = 10.18SPDD134 pKa = 3.16VTALGDD140 pKa = 3.5GGFVISYY147 pKa = 10.05DD148 pKa = 3.64HH149 pKa = 7.03LFGSTDD155 pKa = 4.01LDD157 pKa = 3.69TQAVVFNADD166 pKa = 3.11GTVRR170 pKa = 11.84TSLAVNVGTYY180 pKa = 7.26YY181 pKa = 10.43TGQSAVAGLTNGNFMVSYY199 pKa = 10.48ARR201 pKa = 11.84EE202 pKa = 3.96RR203 pKa = 11.84QDD205 pKa = 3.85GSGATDD211 pKa = 3.63LAFKK215 pKa = 10.49IFNANGGVVRR225 pKa = 11.84AEE227 pKa = 4.26TLADD231 pKa = 3.34TYY233 pKa = 11.35GTMKK237 pKa = 10.53VPGEE241 pKa = 4.28AIGLKK246 pKa = 10.5DD247 pKa = 3.28GGFAIVYY254 pKa = 10.0FDD256 pKa = 5.27DD257 pKa = 3.8GWTGRR262 pKa = 11.84PAEE265 pKa = 4.2VTLGIWNADD274 pKa = 2.87GSLRR278 pKa = 11.84TIQRR282 pKa = 11.84ASEE285 pKa = 3.81PLTRR289 pKa = 11.84GGQRR293 pKa = 11.84NDD295 pKa = 3.2PTIAQLDD302 pKa = 3.8NGFLAVGWSNVTDD315 pKa = 3.48VTTEE319 pKa = 3.9YY320 pKa = 10.94AIWTPDD326 pKa = 2.92GTAVTSGLWRR336 pKa = 11.84GTSQNLNFAAAANGVLSGIVTDD358 pKa = 4.67TIGDD362 pKa = 3.63GSGYY366 pKa = 10.6GIASNTLAFTRR377 pKa = 11.84DD378 pKa = 3.22TRR380 pKa = 11.84GDD382 pKa = 3.49ASSEE386 pKa = 4.2AITGDD391 pKa = 3.56SLRR394 pKa = 11.84DD395 pKa = 3.57FMMGGGGNDD404 pKa = 3.16TLLGYY409 pKa = 10.39GADD412 pKa = 3.78DD413 pKa = 4.01TLDD416 pKa = 3.63GGIGNDD422 pKa = 3.86SVDD425 pKa = 3.38GGIGNDD431 pKa = 3.84LVSGGAGADD440 pKa = 3.26ILIGGTGNDD449 pKa = 3.65TLDD452 pKa = 3.84GGSEE456 pKa = 3.96NDD458 pKa = 3.87RR459 pKa = 11.84LDD461 pKa = 3.86GGSGDD466 pKa = 4.56DD467 pKa = 4.66LLTGGLGADD476 pKa = 3.57ALLGRR481 pKa = 11.84DD482 pKa = 3.77GFDD485 pKa = 2.99TASYY489 pKa = 9.95AGAVASLTADD499 pKa = 4.06LLTPANNTGEE509 pKa = 4.26AAGDD513 pKa = 3.71SYY515 pKa = 12.04AGIEE519 pKa = 4.11GLLGSRR525 pKa = 11.84YY526 pKa = 9.45SDD528 pKa = 3.06RR529 pKa = 11.84LTGNATANRR538 pKa = 11.84LEE540 pKa = 4.51GGAGNDD546 pKa = 3.93TLDD549 pKa = 3.94GGAGNDD555 pKa = 3.76TLVGGDD561 pKa = 3.6GADD564 pKa = 3.03WLYY567 pKa = 11.71ASAGYY572 pKa = 10.48DD573 pKa = 3.79LLTGGAGGDD582 pKa = 2.98RR583 pKa = 11.84FAFTSVSPQYY593 pKa = 10.21DD594 pKa = 3.66TIGDD598 pKa = 3.88FVHH601 pKa = 6.73AQGDD605 pKa = 4.34RR606 pKa = 11.84IVLTSSAFGGLTGLVAGKK624 pKa = 10.37SFVASGSPAAAQAGPTMLYY643 pKa = 8.68NTGNGVLSYY652 pKa = 10.98DD653 pKa = 4.09ADD655 pKa = 3.87GTGAGAATQIATLSGAPTLVFWDD678 pKa = 4.87FVFVV682 pKa = 3.6
MM1 pKa = 7.5ATTSYY6 pKa = 11.18YY7 pKa = 10.49FGYY10 pKa = 10.64LNRR13 pKa = 11.84NEE15 pKa = 4.37PLNTYY20 pKa = 9.27TYY22 pKa = 10.44LSQYY26 pKa = 10.83ADD28 pKa = 3.9TITALNNGGFFTAYY42 pKa = 9.47EE43 pKa = 4.57GYY45 pKa = 10.82YY46 pKa = 10.6DD47 pKa = 4.1GGYY50 pKa = 10.79NYY52 pKa = 10.19ILARR56 pKa = 11.84GLDD59 pKa = 3.75ATGAPAPKK67 pKa = 10.64GEE69 pKa = 4.26FAATNYY75 pKa = 10.49GPGINVRR82 pKa = 11.84SAEE85 pKa = 3.95AATLKK90 pKa = 10.7NGNVVMTYY98 pKa = 7.04TTQSSTSTPITFRR111 pKa = 11.84IFSPDD116 pKa = 2.91GTLLTEE122 pKa = 4.96GSTASGSNYY131 pKa = 10.18SPDD134 pKa = 3.16VTALGDD140 pKa = 3.5GGFVISYY147 pKa = 10.05DD148 pKa = 3.64HH149 pKa = 7.03LFGSTDD155 pKa = 4.01LDD157 pKa = 3.69TQAVVFNADD166 pKa = 3.11GTVRR170 pKa = 11.84TSLAVNVGTYY180 pKa = 7.26YY181 pKa = 10.43TGQSAVAGLTNGNFMVSYY199 pKa = 10.48ARR201 pKa = 11.84EE202 pKa = 3.96RR203 pKa = 11.84QDD205 pKa = 3.85GSGATDD211 pKa = 3.63LAFKK215 pKa = 10.49IFNANGGVVRR225 pKa = 11.84AEE227 pKa = 4.26TLADD231 pKa = 3.34TYY233 pKa = 11.35GTMKK237 pKa = 10.53VPGEE241 pKa = 4.28AIGLKK246 pKa = 10.5DD247 pKa = 3.28GGFAIVYY254 pKa = 10.0FDD256 pKa = 5.27DD257 pKa = 3.8GWTGRR262 pKa = 11.84PAEE265 pKa = 4.2VTLGIWNADD274 pKa = 2.87GSLRR278 pKa = 11.84TIQRR282 pKa = 11.84ASEE285 pKa = 3.81PLTRR289 pKa = 11.84GGQRR293 pKa = 11.84NDD295 pKa = 3.2PTIAQLDD302 pKa = 3.8NGFLAVGWSNVTDD315 pKa = 3.48VTTEE319 pKa = 3.9YY320 pKa = 10.94AIWTPDD326 pKa = 2.92GTAVTSGLWRR336 pKa = 11.84GTSQNLNFAAAANGVLSGIVTDD358 pKa = 4.67TIGDD362 pKa = 3.63GSGYY366 pKa = 10.6GIASNTLAFTRR377 pKa = 11.84DD378 pKa = 3.22TRR380 pKa = 11.84GDD382 pKa = 3.49ASSEE386 pKa = 4.2AITGDD391 pKa = 3.56SLRR394 pKa = 11.84DD395 pKa = 3.57FMMGGGGNDD404 pKa = 3.16TLLGYY409 pKa = 10.39GADD412 pKa = 3.78DD413 pKa = 4.01TLDD416 pKa = 3.63GGIGNDD422 pKa = 3.86SVDD425 pKa = 3.38GGIGNDD431 pKa = 3.84LVSGGAGADD440 pKa = 3.26ILIGGTGNDD449 pKa = 3.65TLDD452 pKa = 3.84GGSEE456 pKa = 3.96NDD458 pKa = 3.87RR459 pKa = 11.84LDD461 pKa = 3.86GGSGDD466 pKa = 4.56DD467 pKa = 4.66LLTGGLGADD476 pKa = 3.57ALLGRR481 pKa = 11.84DD482 pKa = 3.77GFDD485 pKa = 2.99TASYY489 pKa = 9.95AGAVASLTADD499 pKa = 4.06LLTPANNTGEE509 pKa = 4.26AAGDD513 pKa = 3.71SYY515 pKa = 12.04AGIEE519 pKa = 4.11GLLGSRR525 pKa = 11.84YY526 pKa = 9.45SDD528 pKa = 3.06RR529 pKa = 11.84LTGNATANRR538 pKa = 11.84LEE540 pKa = 4.51GGAGNDD546 pKa = 3.93TLDD549 pKa = 3.94GGAGNDD555 pKa = 3.76TLVGGDD561 pKa = 3.6GADD564 pKa = 3.03WLYY567 pKa = 11.71ASAGYY572 pKa = 10.48DD573 pKa = 3.79LLTGGAGGDD582 pKa = 2.98RR583 pKa = 11.84FAFTSVSPQYY593 pKa = 10.21DD594 pKa = 3.66TIGDD598 pKa = 3.88FVHH601 pKa = 6.73AQGDD605 pKa = 4.34RR606 pKa = 11.84IVLTSSAFGGLTGLVAGKK624 pKa = 10.37SFVASGSPAAAQAGPTMLYY643 pKa = 8.68NTGNGVLSYY652 pKa = 10.98DD653 pKa = 4.09ADD655 pKa = 3.87GTGAGAATQIATLSGAPTLVFWDD678 pKa = 4.87FVFVV682 pKa = 3.6
Molecular weight: 69.37 kDa
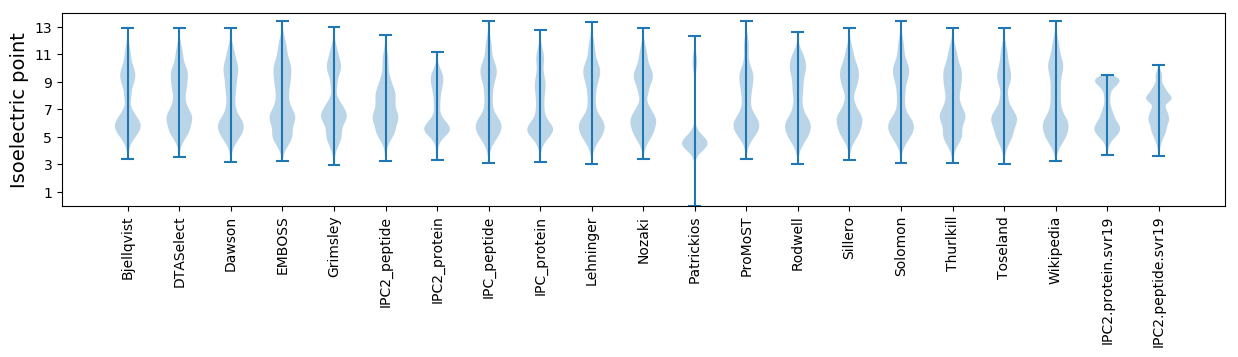
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S2V3S6|A0A3S2V3S6_9RHIZ ABC transporter ATP-binding protein OS=Methylobacterium sp. TER-1 OX=2499852 GN=EOE48_21900 PE=3 SV=1
RR1 pKa = 7.52RR2 pKa = 11.84RR3 pKa = 11.84PHH5 pKa = 6.35PGRR8 pKa = 11.84RR9 pKa = 11.84PAHH12 pKa = 6.09PRR14 pKa = 11.84PPAHH18 pKa = 6.47RR19 pKa = 11.84PHH21 pKa = 6.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84PRR26 pKa = 11.84VVFPHH31 pKa = 6.06RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84HH36 pKa = 4.58RR37 pKa = 11.84PARR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84VRR44 pKa = 11.84PRR46 pKa = 11.84RR47 pKa = 11.84ARR49 pKa = 11.84RR50 pKa = 11.84PRR52 pKa = 11.84RR53 pKa = 11.84APARR57 pKa = 11.84PGLAPLSRR65 pKa = 11.84QGVRR69 pKa = 11.84PEE71 pKa = 3.92FQKK74 pKa = 11.32LEE76 pKa = 3.85AALAVTQPP84 pKa = 3.42
RR1 pKa = 7.52RR2 pKa = 11.84RR3 pKa = 11.84PHH5 pKa = 6.35PGRR8 pKa = 11.84RR9 pKa = 11.84PAHH12 pKa = 6.09PRR14 pKa = 11.84PPAHH18 pKa = 6.47RR19 pKa = 11.84PHH21 pKa = 6.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84PRR26 pKa = 11.84VVFPHH31 pKa = 6.06RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84HH36 pKa = 4.58RR37 pKa = 11.84PARR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84VRR44 pKa = 11.84PRR46 pKa = 11.84RR47 pKa = 11.84ARR49 pKa = 11.84RR50 pKa = 11.84PRR52 pKa = 11.84RR53 pKa = 11.84APARR57 pKa = 11.84PGLAPLSRR65 pKa = 11.84QGVRR69 pKa = 11.84PEE71 pKa = 3.92FQKK74 pKa = 11.32LEE76 pKa = 3.85AALAVTQPP84 pKa = 3.42
Molecular weight: 10.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1810136 |
29 |
8416 |
321.8 |
34.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.717 ± 0.061 | 0.776 ± 0.011 |
5.679 ± 0.03 | 5.28 ± 0.037 |
3.302 ± 0.019 | 9.591 ± 0.053 |
1.912 ± 0.019 | 4.042 ± 0.024 |
2.093 ± 0.027 | 10.62 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.931 ± 0.017 | 1.971 ± 0.021 |
6.005 ± 0.031 | 2.602 ± 0.017 |
8.485 ± 0.046 | 4.64 ± 0.029 |
5.282 ± 0.048 | 7.851 ± 0.024 |
1.225 ± 0.013 | 1.997 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
