
Colletotrichum orchidophilum
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Glomerellales; Glomerellaceae; Colletotrichum
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
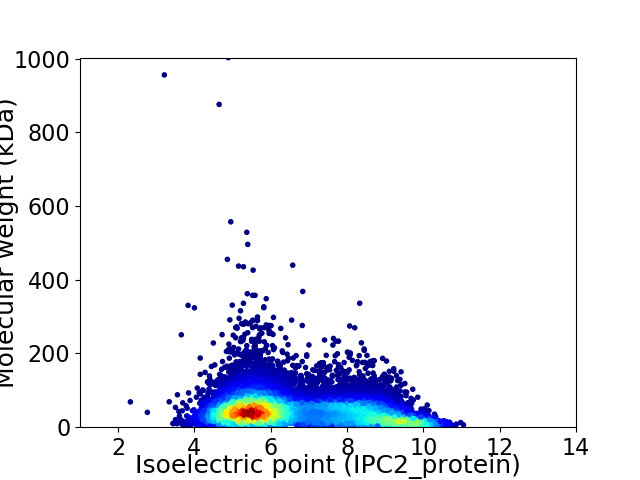
Virtual 2D-PAGE plot for 14495 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G4BEV4|A0A1G4BEV4_9PEZI Cation transporter OS=Colletotrichum orchidophilum OX=1209926 GN=CORC01_04852 PE=4 SV=1
MM1 pKa = 7.49PGLWKK6 pKa = 10.67LLLPLLLLGGRR17 pKa = 11.84SLAQTCLDD25 pKa = 4.17ALNNDD30 pKa = 3.25QVDD33 pKa = 3.51ACVCYY38 pKa = 10.17SLEE41 pKa = 4.19YY42 pKa = 9.39VVSYY46 pKa = 9.97VAYY49 pKa = 8.72IAVSTSYY56 pKa = 10.76TSNCEE61 pKa = 3.91ALPTLSATTSAAARR75 pKa = 11.84FLEE78 pKa = 4.44SNLGGGNYY86 pKa = 9.73SGYY89 pKa = 8.68VTISNGVYY97 pKa = 9.65TGTDD101 pKa = 2.77ILAGPTTRR109 pKa = 11.84AILGLDD115 pKa = 3.42GSGTLVVEE123 pKa = 4.96DD124 pKa = 4.45PPSYY128 pKa = 8.34TTVSRR133 pKa = 11.84TGTATVSPVSLPSFINSTSFLTTNPITAPQTILVANPTPGASIASRR179 pKa = 11.84GTVVVAVPDD188 pKa = 3.63PRR190 pKa = 11.84LYY192 pKa = 9.77NTSTLAYY199 pKa = 10.16AGTDD203 pKa = 3.36PLGAARR209 pKa = 11.84TTISTIAEE217 pKa = 4.12SGFVPGTYY225 pKa = 9.8VVAVATTTVPDD236 pKa = 3.15VSYY239 pKa = 11.13DD240 pKa = 3.54VSTVAYY246 pKa = 8.22TGTAALSGPTVVSVIQPTNRR266 pKa = 11.84SGRR269 pKa = 11.84GTVVIGVPPTGVPPPQVTSIVPYY292 pKa = 9.86EE293 pKa = 4.26GTSSITGATTILNRR307 pKa = 11.84PFDD310 pKa = 3.78GTVPGTVVVGLPPSSFITQTVPFTGKK336 pKa = 9.9EE337 pKa = 4.01VLTAPITISIIPPSGSQLGTVVVGSPSATFLASFVPFTGSTQEE380 pKa = 4.21TLSVIQPTGTSPGIVIIGQPGDD402 pKa = 3.56PYY404 pKa = 11.76ADD406 pKa = 3.63VNDD409 pKa = 4.04PTLTSASQVMPTRR422 pKa = 11.84YY423 pKa = 7.86VTSIVTYY430 pKa = 9.33TPASDD435 pKa = 4.29APNTPSTLAVIDD447 pKa = 3.66GTVIIGLPRR456 pKa = 11.84IISPAATGSDD466 pKa = 3.45SFITSARR473 pKa = 11.84GSFVTSTIDD482 pKa = 3.4FTEE485 pKa = 4.78GADD488 pKa = 3.75LPTGVDD494 pKa = 3.57PVGTIPGTVFVYY506 pKa = 10.67APSAARR512 pKa = 11.84DD513 pKa = 3.68LGPSFVTSEE522 pKa = 4.25VILLGTAAGSAPTTVSIVEE541 pKa = 4.3PTDD544 pKa = 3.65GLPGTIIIGRR554 pKa = 11.84PAIFVTSTVPWTGSIPLPSRR574 pKa = 11.84TTVAVVAPSGDD585 pKa = 3.62SPGTIFVADD594 pKa = 3.62PTSFGRR600 pKa = 11.84QFSTSFVRR608 pKa = 11.84LSGSLEE614 pKa = 3.77EE615 pKa = 4.78GAATTLTLIPGQGTSQPVVVIGVPDD640 pKa = 4.29DD641 pKa = 4.79YY642 pKa = 10.9ITSTVDD648 pKa = 3.53FSGTGILTTRR658 pKa = 11.84TTLSTLPPNGVAPGVIIVADD678 pKa = 4.23PPTSVSPGSAVVTAGVSDD696 pKa = 4.22GTQPNYY702 pKa = 9.17VTSTVVYY709 pKa = 10.08GGLTTSVLTSVPPSGSNRR727 pKa = 11.84GTVVLAVPSAAAGNPSPTPVSQLNNPSVARR757 pKa = 11.84ASFVTSTIVSLVGSLVASRR776 pKa = 11.84ATISTVTPDD785 pKa = 3.26IANSIPGTYY794 pKa = 9.75VVALPNAYY802 pKa = 7.99ITSTVPWTGEE812 pKa = 4.05VPLSTVTTISIIPPSNFQPGVIVVAAPNAIDD843 pKa = 3.85SSTTQAVGSGTVPTPVLASNASPGNSPTPLSVTQLPAQTLFTTSTVRR890 pKa = 11.84LASDD894 pKa = 4.17LGISSATILTTQQASPGSPSTVLLGVPDD922 pKa = 3.52QVANSEE928 pKa = 4.08ASTAGAEE935 pKa = 4.22AEE937 pKa = 4.5TIINPNSGPQPASSSAAVFTSTATEE962 pKa = 3.83SSIAFSASVGSVPSTTVLGNTALSSFVISDD992 pKa = 3.39IFYY995 pKa = 9.41TGTNSFTSPIPLTTLSADD1013 pKa = 3.6LGSPMTVILLQPSSAIGSTGNPVGGSGSTSSLATGSSPTTSASPFSNVAGTAFSTIASTSVLADD1077 pKa = 3.28SSISTTASSEE1087 pKa = 4.07RR1088 pKa = 11.84FSNQSPFGTPPDD1100 pKa = 3.9SAVTTPLATVFVDD1113 pKa = 3.49YY1114 pKa = 10.43TGTGVPSEE1122 pKa = 4.2FLTLRR1127 pKa = 11.84TTSGTVVIQRR1137 pKa = 11.84PRR1139 pKa = 11.84PLTTVQTFYY1148 pKa = 11.01SGSVSIAILTIIPSGTEE1165 pKa = 2.99AATIVFGVPSPVNTTPVTSFSQEE1188 pKa = 4.08SLSVEE1193 pKa = 4.35LPSSTNSIDD1202 pKa = 3.59TLISADD1208 pKa = 3.49PQIIVSSTARR1218 pKa = 11.84SSTPVAVPGTSTGTNQTSSEE1238 pKa = 4.14LVADD1242 pKa = 3.92PTSQISSPISSTRR1255 pKa = 11.84SSSSEE1260 pKa = 4.02TISSSNNSGAGSPALSAILSRR1281 pKa = 11.84VSSTLTSLDD1290 pKa = 3.31STIEE1294 pKa = 4.04SSSSVLAQPQATSLSSLTASSTANTATATTVTDD1327 pKa = 3.69AALTSNSVSEE1337 pKa = 4.49SSSSSFSPILSGSVAPTASSSQSTSSGAISLSIISLDD1374 pKa = 3.48VSTSDD1379 pKa = 4.37PSSQGSGTFTSNTPPPSLLSTSISITADD1407 pKa = 3.18PNLISSILSTATSLSTSDD1425 pKa = 3.57AAFSASSGGLSLSATVEE1442 pKa = 4.01VSVEE1446 pKa = 4.17STVSSTAARR1455 pKa = 11.84TPSTPSTSPSTSLSSVLDD1473 pKa = 3.49ILSDD1477 pKa = 3.51TTASSFSSSSGSGSQSSTSSDD1498 pKa = 2.88ATASTSPSSEE1508 pKa = 3.87LSIQNGPSSNLASTDD1523 pKa = 3.14VPAPSDD1529 pKa = 3.78SVVSSTALPNILSSLSEE1546 pKa = 4.44SIEE1549 pKa = 4.11SSSTLISSSSGTSTSTGTDD1568 pKa = 2.85STTAEE1573 pKa = 4.18TNAVSNDD1580 pKa = 2.97SSAQSMNPVSSNPSDD1595 pKa = 3.54SSTSSLDD1602 pKa = 3.17PGISVGSSGLIGSTTTSGLSLVTPTATVVTSVVVFTGTEE1641 pKa = 4.15SFTGTGLVPLTTIYY1655 pKa = 10.26PSNSEE1660 pKa = 3.82APTVVFGAARR1670 pKa = 11.84EE1671 pKa = 4.29LFSSVAVPYY1680 pKa = 10.29SGTGIITGPQQVSIVLPSGSDD1701 pKa = 3.36SLVLGTIVYY1710 pKa = 7.71ATPDD1714 pKa = 3.58PFVTSEE1720 pKa = 3.5VDD1722 pKa = 3.35YY1723 pKa = 10.53TGTGTLVSPLPLTTIYY1739 pKa = 10.75QSGTVPGTVILARR1752 pKa = 11.84PAIFATSSTSSSTHH1766 pKa = 4.52STASSEE1772 pKa = 4.15SSISIEE1778 pKa = 4.8LSTTSQSRR1786 pKa = 11.84SSSDD1790 pKa = 3.18SNASLEE1796 pKa = 4.4TSSSFASSSQLEE1808 pKa = 4.36STPSSTSSEE1817 pKa = 4.27SLLGPSNIPEE1827 pKa = 4.41STTSSTPTVLAEE1839 pKa = 4.22SGTASEE1845 pKa = 4.5SQSPSSEE1852 pKa = 4.11LASTAPSTTSEE1863 pKa = 4.48GVSSTIGEE1871 pKa = 4.34LSSSTIISASSIGSSSSLGPSSSIAALTDD1900 pKa = 3.53ASLSTGVSPDD1910 pKa = 3.44PAFLSTSPISISDD1923 pKa = 3.64STSPTLTAEE1932 pKa = 4.22SSISSSRR1939 pKa = 11.84TTLSTVPDD1947 pKa = 3.73TTTSQRR1953 pKa = 11.84SPFSTEE1959 pKa = 3.44ISSSSQAPEE1968 pKa = 3.65ISSQQSSDD1976 pKa = 3.27FTALSSSTASSTTPLPAFSSTTEE1999 pKa = 3.83SSSIEE2004 pKa = 4.06AATSPSTADD2013 pKa = 3.02ASMTSSSSSTDD2024 pKa = 3.05STGMLEE2030 pKa = 3.95ATTGFEE2036 pKa = 4.26ATSGATSSSGVEE2048 pKa = 4.1SGSATDD2054 pKa = 3.34TGTTGAAFASLLTTSGSSSSSDD2076 pKa = 2.69IAATSNIADD2085 pKa = 3.92PRR2087 pKa = 11.84SSSTLSDD2094 pKa = 3.51VLSSEE2099 pKa = 4.5TSVSTSISIEE2109 pKa = 4.13SEE2111 pKa = 3.86ASSATSMDD2119 pKa = 3.27TTTTEE2124 pKa = 4.36LTSSTSTIASSLAIEE2139 pKa = 4.58SLTTPIVTTSEE2150 pKa = 4.11TTSNDD2155 pKa = 2.25ISRR2158 pKa = 11.84SISTTSITSEE2168 pKa = 4.03SDD2170 pKa = 3.27QSSSTSDD2177 pKa = 2.9IEE2179 pKa = 4.62TVSTSYY2185 pKa = 11.56ASLIPTTDD2193 pKa = 3.06ASSLSGISITAGGTTSEE2210 pKa = 4.54PSSSVADD2217 pKa = 3.38ITSLISSTDD2226 pKa = 3.31IPPTTPTSLFSEE2238 pKa = 4.51ATSTSSEE2245 pKa = 4.08ATTSTGDD2252 pKa = 3.61PPSTPSSSLADD2263 pKa = 3.56AASSLSLTDD2272 pKa = 3.71ADD2274 pKa = 4.41TVTSSSLLPTSSGLATATPISSEE2297 pKa = 4.1TSPVSITTASVDD2309 pKa = 3.07AASYY2313 pKa = 8.1VTSQLIYY2320 pKa = 9.48TGPSSLSGPLPLSTIQPTNGSPGTVILAFPASFYY2354 pKa = 7.6TTSTSIYY2361 pKa = 9.55TEE2363 pKa = 4.09GDD2365 pKa = 3.53SYY2367 pKa = 11.26IVPSAISTVAPSGIAKK2383 pKa = 8.36GTVIFAARR2391 pKa = 11.84YY2392 pKa = 7.76AVARR2396 pKa = 11.84VVYY2399 pKa = 10.05SGTSVITEE2407 pKa = 5.29AIPVATYY2414 pKa = 9.43PGSGTIIGTIVYY2426 pKa = 9.15AYY2428 pKa = 7.57PPPSLATSEE2437 pKa = 4.48EE2438 pKa = 4.57STSGASTTASTEE2450 pKa = 4.06PLSSITPQVNSTSSTSSDD2468 pKa = 3.43VNFSSTPSSSSTLSTFNLWIDD2489 pKa = 3.96DD2490 pKa = 3.93RR2491 pKa = 11.84GADD2494 pKa = 3.38NVEE2497 pKa = 3.68RR2498 pKa = 11.84ATFRR2502 pKa = 11.84DD2503 pKa = 3.93YY2504 pKa = 10.91IITIDD2509 pKa = 3.5RR2510 pKa = 11.84FYY2512 pKa = 11.66GLLSFIDD2519 pKa = 4.12FVYY2522 pKa = 10.94SS2523 pKa = 3.39
MM1 pKa = 7.49PGLWKK6 pKa = 10.67LLLPLLLLGGRR17 pKa = 11.84SLAQTCLDD25 pKa = 4.17ALNNDD30 pKa = 3.25QVDD33 pKa = 3.51ACVCYY38 pKa = 10.17SLEE41 pKa = 4.19YY42 pKa = 9.39VVSYY46 pKa = 9.97VAYY49 pKa = 8.72IAVSTSYY56 pKa = 10.76TSNCEE61 pKa = 3.91ALPTLSATTSAAARR75 pKa = 11.84FLEE78 pKa = 4.44SNLGGGNYY86 pKa = 9.73SGYY89 pKa = 8.68VTISNGVYY97 pKa = 9.65TGTDD101 pKa = 2.77ILAGPTTRR109 pKa = 11.84AILGLDD115 pKa = 3.42GSGTLVVEE123 pKa = 4.96DD124 pKa = 4.45PPSYY128 pKa = 8.34TTVSRR133 pKa = 11.84TGTATVSPVSLPSFINSTSFLTTNPITAPQTILVANPTPGASIASRR179 pKa = 11.84GTVVVAVPDD188 pKa = 3.63PRR190 pKa = 11.84LYY192 pKa = 9.77NTSTLAYY199 pKa = 10.16AGTDD203 pKa = 3.36PLGAARR209 pKa = 11.84TTISTIAEE217 pKa = 4.12SGFVPGTYY225 pKa = 9.8VVAVATTTVPDD236 pKa = 3.15VSYY239 pKa = 11.13DD240 pKa = 3.54VSTVAYY246 pKa = 8.22TGTAALSGPTVVSVIQPTNRR266 pKa = 11.84SGRR269 pKa = 11.84GTVVIGVPPTGVPPPQVTSIVPYY292 pKa = 9.86EE293 pKa = 4.26GTSSITGATTILNRR307 pKa = 11.84PFDD310 pKa = 3.78GTVPGTVVVGLPPSSFITQTVPFTGKK336 pKa = 9.9EE337 pKa = 4.01VLTAPITISIIPPSGSQLGTVVVGSPSATFLASFVPFTGSTQEE380 pKa = 4.21TLSVIQPTGTSPGIVIIGQPGDD402 pKa = 3.56PYY404 pKa = 11.76ADD406 pKa = 3.63VNDD409 pKa = 4.04PTLTSASQVMPTRR422 pKa = 11.84YY423 pKa = 7.86VTSIVTYY430 pKa = 9.33TPASDD435 pKa = 4.29APNTPSTLAVIDD447 pKa = 3.66GTVIIGLPRR456 pKa = 11.84IISPAATGSDD466 pKa = 3.45SFITSARR473 pKa = 11.84GSFVTSTIDD482 pKa = 3.4FTEE485 pKa = 4.78GADD488 pKa = 3.75LPTGVDD494 pKa = 3.57PVGTIPGTVFVYY506 pKa = 10.67APSAARR512 pKa = 11.84DD513 pKa = 3.68LGPSFVTSEE522 pKa = 4.25VILLGTAAGSAPTTVSIVEE541 pKa = 4.3PTDD544 pKa = 3.65GLPGTIIIGRR554 pKa = 11.84PAIFVTSTVPWTGSIPLPSRR574 pKa = 11.84TTVAVVAPSGDD585 pKa = 3.62SPGTIFVADD594 pKa = 3.62PTSFGRR600 pKa = 11.84QFSTSFVRR608 pKa = 11.84LSGSLEE614 pKa = 3.77EE615 pKa = 4.78GAATTLTLIPGQGTSQPVVVIGVPDD640 pKa = 4.29DD641 pKa = 4.79YY642 pKa = 10.9ITSTVDD648 pKa = 3.53FSGTGILTTRR658 pKa = 11.84TTLSTLPPNGVAPGVIIVADD678 pKa = 4.23PPTSVSPGSAVVTAGVSDD696 pKa = 4.22GTQPNYY702 pKa = 9.17VTSTVVYY709 pKa = 10.08GGLTTSVLTSVPPSGSNRR727 pKa = 11.84GTVVLAVPSAAAGNPSPTPVSQLNNPSVARR757 pKa = 11.84ASFVTSTIVSLVGSLVASRR776 pKa = 11.84ATISTVTPDD785 pKa = 3.26IANSIPGTYY794 pKa = 9.75VVALPNAYY802 pKa = 7.99ITSTVPWTGEE812 pKa = 4.05VPLSTVTTISIIPPSNFQPGVIVVAAPNAIDD843 pKa = 3.85SSTTQAVGSGTVPTPVLASNASPGNSPTPLSVTQLPAQTLFTTSTVRR890 pKa = 11.84LASDD894 pKa = 4.17LGISSATILTTQQASPGSPSTVLLGVPDD922 pKa = 3.52QVANSEE928 pKa = 4.08ASTAGAEE935 pKa = 4.22AEE937 pKa = 4.5TIINPNSGPQPASSSAAVFTSTATEE962 pKa = 3.83SSIAFSASVGSVPSTTVLGNTALSSFVISDD992 pKa = 3.39IFYY995 pKa = 9.41TGTNSFTSPIPLTTLSADD1013 pKa = 3.6LGSPMTVILLQPSSAIGSTGNPVGGSGSTSSLATGSSPTTSASPFSNVAGTAFSTIASTSVLADD1077 pKa = 3.28SSISTTASSEE1087 pKa = 4.07RR1088 pKa = 11.84FSNQSPFGTPPDD1100 pKa = 3.9SAVTTPLATVFVDD1113 pKa = 3.49YY1114 pKa = 10.43TGTGVPSEE1122 pKa = 4.2FLTLRR1127 pKa = 11.84TTSGTVVIQRR1137 pKa = 11.84PRR1139 pKa = 11.84PLTTVQTFYY1148 pKa = 11.01SGSVSIAILTIIPSGTEE1165 pKa = 2.99AATIVFGVPSPVNTTPVTSFSQEE1188 pKa = 4.08SLSVEE1193 pKa = 4.35LPSSTNSIDD1202 pKa = 3.59TLISADD1208 pKa = 3.49PQIIVSSTARR1218 pKa = 11.84SSTPVAVPGTSTGTNQTSSEE1238 pKa = 4.14LVADD1242 pKa = 3.92PTSQISSPISSTRR1255 pKa = 11.84SSSSEE1260 pKa = 4.02TISSSNNSGAGSPALSAILSRR1281 pKa = 11.84VSSTLTSLDD1290 pKa = 3.31STIEE1294 pKa = 4.04SSSSVLAQPQATSLSSLTASSTANTATATTVTDD1327 pKa = 3.69AALTSNSVSEE1337 pKa = 4.49SSSSSFSPILSGSVAPTASSSQSTSSGAISLSIISLDD1374 pKa = 3.48VSTSDD1379 pKa = 4.37PSSQGSGTFTSNTPPPSLLSTSISITADD1407 pKa = 3.18PNLISSILSTATSLSTSDD1425 pKa = 3.57AAFSASSGGLSLSATVEE1442 pKa = 4.01VSVEE1446 pKa = 4.17STVSSTAARR1455 pKa = 11.84TPSTPSTSPSTSLSSVLDD1473 pKa = 3.49ILSDD1477 pKa = 3.51TTASSFSSSSGSGSQSSTSSDD1498 pKa = 2.88ATASTSPSSEE1508 pKa = 3.87LSIQNGPSSNLASTDD1523 pKa = 3.14VPAPSDD1529 pKa = 3.78SVVSSTALPNILSSLSEE1546 pKa = 4.44SIEE1549 pKa = 4.11SSSTLISSSSGTSTSTGTDD1568 pKa = 2.85STTAEE1573 pKa = 4.18TNAVSNDD1580 pKa = 2.97SSAQSMNPVSSNPSDD1595 pKa = 3.54SSTSSLDD1602 pKa = 3.17PGISVGSSGLIGSTTTSGLSLVTPTATVVTSVVVFTGTEE1641 pKa = 4.15SFTGTGLVPLTTIYY1655 pKa = 10.26PSNSEE1660 pKa = 3.82APTVVFGAARR1670 pKa = 11.84EE1671 pKa = 4.29LFSSVAVPYY1680 pKa = 10.29SGTGIITGPQQVSIVLPSGSDD1701 pKa = 3.36SLVLGTIVYY1710 pKa = 7.71ATPDD1714 pKa = 3.58PFVTSEE1720 pKa = 3.5VDD1722 pKa = 3.35YY1723 pKa = 10.53TGTGTLVSPLPLTTIYY1739 pKa = 10.75QSGTVPGTVILARR1752 pKa = 11.84PAIFATSSTSSSTHH1766 pKa = 4.52STASSEE1772 pKa = 4.15SSISIEE1778 pKa = 4.8LSTTSQSRR1786 pKa = 11.84SSSDD1790 pKa = 3.18SNASLEE1796 pKa = 4.4TSSSFASSSQLEE1808 pKa = 4.36STPSSTSSEE1817 pKa = 4.27SLLGPSNIPEE1827 pKa = 4.41STTSSTPTVLAEE1839 pKa = 4.22SGTASEE1845 pKa = 4.5SQSPSSEE1852 pKa = 4.11LASTAPSTTSEE1863 pKa = 4.48GVSSTIGEE1871 pKa = 4.34LSSSTIISASSIGSSSSLGPSSSIAALTDD1900 pKa = 3.53ASLSTGVSPDD1910 pKa = 3.44PAFLSTSPISISDD1923 pKa = 3.64STSPTLTAEE1932 pKa = 4.22SSISSSRR1939 pKa = 11.84TTLSTVPDD1947 pKa = 3.73TTTSQRR1953 pKa = 11.84SPFSTEE1959 pKa = 3.44ISSSSQAPEE1968 pKa = 3.65ISSQQSSDD1976 pKa = 3.27FTALSSSTASSTTPLPAFSSTTEE1999 pKa = 3.83SSSIEE2004 pKa = 4.06AATSPSTADD2013 pKa = 3.02ASMTSSSSSTDD2024 pKa = 3.05STGMLEE2030 pKa = 3.95ATTGFEE2036 pKa = 4.26ATSGATSSSGVEE2048 pKa = 4.1SGSATDD2054 pKa = 3.34TGTTGAAFASLLTTSGSSSSSDD2076 pKa = 2.69IAATSNIADD2085 pKa = 3.92PRR2087 pKa = 11.84SSSTLSDD2094 pKa = 3.51VLSSEE2099 pKa = 4.5TSVSTSISIEE2109 pKa = 4.13SEE2111 pKa = 3.86ASSATSMDD2119 pKa = 3.27TTTTEE2124 pKa = 4.36LTSSTSTIASSLAIEE2139 pKa = 4.58SLTTPIVTTSEE2150 pKa = 4.11TTSNDD2155 pKa = 2.25ISRR2158 pKa = 11.84SISTTSITSEE2168 pKa = 4.03SDD2170 pKa = 3.27QSSSTSDD2177 pKa = 2.9IEE2179 pKa = 4.62TVSTSYY2185 pKa = 11.56ASLIPTTDD2193 pKa = 3.06ASSLSGISITAGGTTSEE2210 pKa = 4.54PSSSVADD2217 pKa = 3.38ITSLISSTDD2226 pKa = 3.31IPPTTPTSLFSEE2238 pKa = 4.51ATSTSSEE2245 pKa = 4.08ATTSTGDD2252 pKa = 3.61PPSTPSSSLADD2263 pKa = 3.56AASSLSLTDD2272 pKa = 3.71ADD2274 pKa = 4.41TVTSSSLLPTSSGLATATPISSEE2297 pKa = 4.1TSPVSITTASVDD2309 pKa = 3.07AASYY2313 pKa = 8.1VTSQLIYY2320 pKa = 9.48TGPSSLSGPLPLSTIQPTNGSPGTVILAFPASFYY2354 pKa = 7.6TTSTSIYY2361 pKa = 9.55TEE2363 pKa = 4.09GDD2365 pKa = 3.53SYY2367 pKa = 11.26IVPSAISTVAPSGIAKK2383 pKa = 8.36GTVIFAARR2391 pKa = 11.84YY2392 pKa = 7.76AVARR2396 pKa = 11.84VVYY2399 pKa = 10.05SGTSVITEE2407 pKa = 5.29AIPVATYY2414 pKa = 9.43PGSGTIIGTIVYY2426 pKa = 9.15AYY2428 pKa = 7.57PPPSLATSEE2437 pKa = 4.48EE2438 pKa = 4.57STSGASTTASTEE2450 pKa = 4.06PLSSITPQVNSTSSTSSDD2468 pKa = 3.43VNFSSTPSSSSTLSTFNLWIDD2489 pKa = 3.96DD2490 pKa = 3.93RR2491 pKa = 11.84GADD2494 pKa = 3.38NVEE2497 pKa = 3.68RR2498 pKa = 11.84ATFRR2502 pKa = 11.84DD2503 pKa = 3.93YY2504 pKa = 10.91IITIDD2509 pKa = 3.5RR2510 pKa = 11.84FYY2512 pKa = 11.66GLLSFIDD2519 pKa = 4.12FVYY2522 pKa = 10.94SS2523 pKa = 3.39
Molecular weight: 250.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G4B2K1|A0A1G4B2K1_9PEZI Uncharacterized protein OS=Colletotrichum orchidophilum OX=1209926 GN=CORC01_09176 PE=4 SV=1
MM1 pKa = 7.85PLRR4 pKa = 11.84STRR7 pKa = 11.84HH8 pKa = 4.05TTTTAPRR15 pKa = 11.84RR16 pKa = 11.84SIFSRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84APAHH27 pKa = 5.9TSHH30 pKa = 6.67HH31 pKa = 5.83HH32 pKa = 5.27TTTKK36 pKa = 7.05TTRR39 pKa = 11.84TTKK42 pKa = 10.47RR43 pKa = 11.84GGGGLFSRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84GPVHH57 pKa = 6.22TAPVHH62 pKa = 4.58HH63 pKa = 6.26QRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 9.81PSMGDD72 pKa = 3.3KK73 pKa = 10.53ISGAMLKK80 pKa = 10.72LKK82 pKa = 9.36GTLTRR87 pKa = 11.84RR88 pKa = 11.84PGQKK92 pKa = 9.9AAGTRR97 pKa = 11.84RR98 pKa = 11.84MHH100 pKa = 5.68GTDD103 pKa = 2.72GRR105 pKa = 11.84GSHH108 pKa = 6.42RR109 pKa = 11.84AHH111 pKa = 7.53RR112 pKa = 11.84YY113 pKa = 8.09
MM1 pKa = 7.85PLRR4 pKa = 11.84STRR7 pKa = 11.84HH8 pKa = 4.05TTTTAPRR15 pKa = 11.84RR16 pKa = 11.84SIFSRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84APAHH27 pKa = 5.9TSHH30 pKa = 6.67HH31 pKa = 5.83HH32 pKa = 5.27TTTKK36 pKa = 7.05TTRR39 pKa = 11.84TTKK42 pKa = 10.47RR43 pKa = 11.84GGGGLFSRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84GPVHH57 pKa = 6.22TAPVHH62 pKa = 4.58HH63 pKa = 6.26QRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 9.81PSMGDD72 pKa = 3.3KK73 pKa = 10.53ISGAMLKK80 pKa = 10.72LKK82 pKa = 9.36GTLTRR87 pKa = 11.84RR88 pKa = 11.84PGQKK92 pKa = 9.9AAGTRR97 pKa = 11.84RR98 pKa = 11.84MHH100 pKa = 5.68GTDD103 pKa = 2.72GRR105 pKa = 11.84GSHH108 pKa = 6.42RR109 pKa = 11.84AHH111 pKa = 7.53RR112 pKa = 11.84YY113 pKa = 8.09
Molecular weight: 12.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6380480 |
21 |
9789 |
440.2 |
48.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.046 ± 0.022 | 1.249 ± 0.008 |
5.678 ± 0.016 | 6.006 ± 0.023 |
3.755 ± 0.012 | 7.108 ± 0.019 |
2.352 ± 0.011 | 4.771 ± 0.015 |
4.803 ± 0.022 | 8.717 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.182 ± 0.009 | 3.629 ± 0.011 |
6.165 ± 0.027 | 3.921 ± 0.013 |
6.052 ± 0.019 | 8.113 ± 0.023 |
6.049 ± 0.027 | 6.23 ± 0.016 |
1.51 ± 0.009 | 2.665 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
