
Pseudospirillum japonicum
Taxonomy: cellular organisms; Bacteria;
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
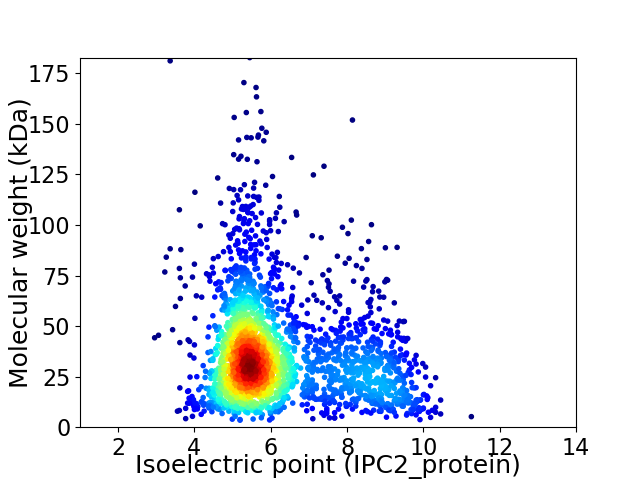
Virtual 2D-PAGE plot for 2385 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
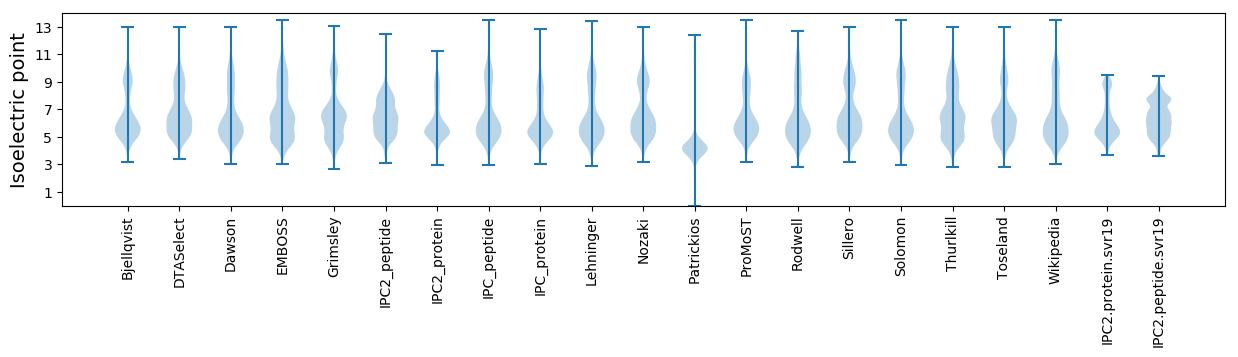
* You can choose from 21 different methods for calculating isoelectric point
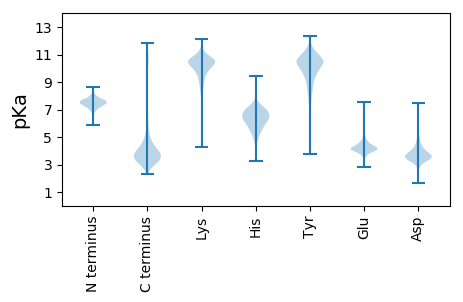
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H6R4W0|A0A1H6R4W0_9GAMM Cytochrome c-type biogenesis protein CcmE OS=Pseudospirillum japonicum OX=64971 GN=ccmE PE=3 SV=1
MM1 pKa = 7.29SVVAQNFTTYY11 pKa = 10.4VYY13 pKa = 11.03NQLLKK18 pKa = 10.67RR19 pKa = 11.84NPDD22 pKa = 3.68AEE24 pKa = 4.1GLTYY28 pKa = 10.05WEE30 pKa = 4.25QQLDD34 pKa = 3.97STQVSAEE41 pKa = 3.87DD42 pKa = 3.28MFVYY46 pKa = 10.41FLEE49 pKa = 4.47SPEE52 pKa = 3.93FSDD55 pKa = 4.19RR56 pKa = 11.84YY57 pKa = 10.42SPIIRR62 pKa = 11.84LYY64 pKa = 8.79MSAFNRR70 pKa = 11.84FPDD73 pKa = 4.06SEE75 pKa = 4.0GMEE78 pKa = 4.04YY79 pKa = 9.75WAQEE83 pKa = 4.04MANGQTPYY91 pKa = 10.8QIARR95 pKa = 11.84LLLSSTEE102 pKa = 3.94YY103 pKa = 10.57TDD105 pKa = 3.65MYY107 pKa = 10.79GSDD110 pKa = 3.41VSHH113 pKa = 6.99SDD115 pKa = 3.58FVNSLYY121 pKa = 11.35VNVLGRR127 pKa = 11.84AGDD130 pKa = 3.9PSGTAYY136 pKa = 9.67WEE138 pKa = 4.12EE139 pKa = 4.48VLTQGLATQKK149 pKa = 10.74DD150 pKa = 4.02VLVSFSEE157 pKa = 4.11SQEE160 pKa = 4.33AIEE163 pKa = 5.16GMAKK167 pKa = 9.9EE168 pKa = 4.41VNTVLAYY175 pKa = 10.68NAMLGRR181 pKa = 11.84EE182 pKa = 4.19PTAEE186 pKa = 4.11EE187 pKa = 4.14LRR189 pKa = 11.84RR190 pKa = 11.84APSDD194 pKa = 3.3PAILVSQLFQGTEE207 pKa = 4.0YY208 pKa = 10.84NGPLVPGYY216 pKa = 9.44EE217 pKa = 3.85LEE219 pKa = 4.35YY220 pKa = 10.48WVDD223 pKa = 3.66TQVLGDD229 pKa = 3.85TLYY232 pKa = 10.27LTDD235 pKa = 4.0KK236 pKa = 10.63SHH238 pKa = 6.6GVVLADD244 pKa = 4.09LRR246 pKa = 11.84EE247 pKa = 4.32DD248 pKa = 3.5QVTDD252 pKa = 2.98AGTVLNVQDD261 pKa = 4.51LVSAVNIDD269 pKa = 3.49ASDD272 pKa = 3.4MRR274 pKa = 11.84FNAARR279 pKa = 11.84LYY281 pKa = 11.31GDD283 pKa = 4.96DD284 pKa = 4.99DD285 pKa = 4.88ANKK288 pKa = 8.92LTATGAADD296 pKa = 3.28ILYY299 pKa = 10.3GRR301 pKa = 11.84AGNDD305 pKa = 3.27LLEE308 pKa = 4.58GLSGNDD314 pKa = 3.16TLYY317 pKa = 11.32GDD319 pKa = 4.23EE320 pKa = 5.35GNDD323 pKa = 3.82TLHH326 pKa = 6.73GGYY329 pKa = 10.69GEE331 pKa = 4.85DD332 pKa = 4.48RR333 pKa = 11.84MFGGDD338 pKa = 3.73GNDD341 pKa = 3.28EE342 pKa = 4.64FIIADD347 pKa = 3.75LDD349 pKa = 4.41EE350 pKa = 4.28IHH352 pKa = 7.09LEE354 pKa = 4.04QEE356 pKa = 4.31EE357 pKa = 4.38LQGGAGTDD365 pKa = 2.91RR366 pKa = 11.84LTLQEE371 pKa = 4.44SGAVDD376 pKa = 4.07LSKK379 pKa = 11.41VLMFDD384 pKa = 3.4MEE386 pKa = 4.42EE387 pKa = 4.73LEE389 pKa = 4.79LSNGGNTVTMVRR401 pKa = 11.84EE402 pKa = 3.92QLLQLTNIVPAAQGDD417 pKa = 4.64SILKK421 pKa = 9.47LAQAATLDD429 pKa = 4.14LEE431 pKa = 5.17QMPQLQAFNKK441 pKa = 9.91IYY443 pKa = 10.68GATTGSNDD451 pKa = 3.38LLGYY455 pKa = 10.78AGDD458 pKa = 4.7DD459 pKa = 3.57YY460 pKa = 11.88LVGGKK465 pKa = 9.44NEE467 pKa = 3.9DD468 pKa = 3.44QLRR471 pKa = 11.84GGEE474 pKa = 4.18GVDD477 pKa = 3.52YY478 pKa = 11.56LEE480 pKa = 5.03GGEE483 pKa = 4.27GADD486 pKa = 2.46IYY488 pKa = 11.47VFNRR492 pKa = 11.84ADD494 pKa = 3.45YY495 pKa = 9.37SQQSVGATDD504 pKa = 4.05FVGDD508 pKa = 4.66TIHH511 pKa = 7.17GLDD514 pKa = 3.74FTQGDD519 pKa = 4.56KK520 pKa = 10.71IRR522 pKa = 11.84LEE524 pKa = 4.34APLSGSTQALSLEE537 pKa = 4.26SGFYY541 pKa = 10.61SSLTEE546 pKa = 4.38TIQANATIQSAFAGDD561 pKa = 4.15DD562 pKa = 3.17VDD564 pKa = 5.73AVLLQVKK571 pKa = 10.02AGTAAGYY578 pKa = 9.36YY579 pKa = 10.54LLVEE583 pKa = 5.08EE584 pKa = 5.11YY585 pKa = 10.81NADD588 pKa = 3.84YY589 pKa = 11.06QPADD593 pKa = 3.62ATDD596 pKa = 3.72TTDD599 pKa = 4.36DD600 pKa = 3.81AASLVNTEE608 pKa = 4.28TDD610 pKa = 3.28TLNYY614 pKa = 10.6DD615 pKa = 3.64PASDD619 pKa = 4.95LIIRR623 pKa = 11.84LTGIEE628 pKa = 5.01SLNQLNGFSTSVFII642 pKa = 5.76
MM1 pKa = 7.29SVVAQNFTTYY11 pKa = 10.4VYY13 pKa = 11.03NQLLKK18 pKa = 10.67RR19 pKa = 11.84NPDD22 pKa = 3.68AEE24 pKa = 4.1GLTYY28 pKa = 10.05WEE30 pKa = 4.25QQLDD34 pKa = 3.97STQVSAEE41 pKa = 3.87DD42 pKa = 3.28MFVYY46 pKa = 10.41FLEE49 pKa = 4.47SPEE52 pKa = 3.93FSDD55 pKa = 4.19RR56 pKa = 11.84YY57 pKa = 10.42SPIIRR62 pKa = 11.84LYY64 pKa = 8.79MSAFNRR70 pKa = 11.84FPDD73 pKa = 4.06SEE75 pKa = 4.0GMEE78 pKa = 4.04YY79 pKa = 9.75WAQEE83 pKa = 4.04MANGQTPYY91 pKa = 10.8QIARR95 pKa = 11.84LLLSSTEE102 pKa = 3.94YY103 pKa = 10.57TDD105 pKa = 3.65MYY107 pKa = 10.79GSDD110 pKa = 3.41VSHH113 pKa = 6.99SDD115 pKa = 3.58FVNSLYY121 pKa = 11.35VNVLGRR127 pKa = 11.84AGDD130 pKa = 3.9PSGTAYY136 pKa = 9.67WEE138 pKa = 4.12EE139 pKa = 4.48VLTQGLATQKK149 pKa = 10.74DD150 pKa = 4.02VLVSFSEE157 pKa = 4.11SQEE160 pKa = 4.33AIEE163 pKa = 5.16GMAKK167 pKa = 9.9EE168 pKa = 4.41VNTVLAYY175 pKa = 10.68NAMLGRR181 pKa = 11.84EE182 pKa = 4.19PTAEE186 pKa = 4.11EE187 pKa = 4.14LRR189 pKa = 11.84RR190 pKa = 11.84APSDD194 pKa = 3.3PAILVSQLFQGTEE207 pKa = 4.0YY208 pKa = 10.84NGPLVPGYY216 pKa = 9.44EE217 pKa = 3.85LEE219 pKa = 4.35YY220 pKa = 10.48WVDD223 pKa = 3.66TQVLGDD229 pKa = 3.85TLYY232 pKa = 10.27LTDD235 pKa = 4.0KK236 pKa = 10.63SHH238 pKa = 6.6GVVLADD244 pKa = 4.09LRR246 pKa = 11.84EE247 pKa = 4.32DD248 pKa = 3.5QVTDD252 pKa = 2.98AGTVLNVQDD261 pKa = 4.51LVSAVNIDD269 pKa = 3.49ASDD272 pKa = 3.4MRR274 pKa = 11.84FNAARR279 pKa = 11.84LYY281 pKa = 11.31GDD283 pKa = 4.96DD284 pKa = 4.99DD285 pKa = 4.88ANKK288 pKa = 8.92LTATGAADD296 pKa = 3.28ILYY299 pKa = 10.3GRR301 pKa = 11.84AGNDD305 pKa = 3.27LLEE308 pKa = 4.58GLSGNDD314 pKa = 3.16TLYY317 pKa = 11.32GDD319 pKa = 4.23EE320 pKa = 5.35GNDD323 pKa = 3.82TLHH326 pKa = 6.73GGYY329 pKa = 10.69GEE331 pKa = 4.85DD332 pKa = 4.48RR333 pKa = 11.84MFGGDD338 pKa = 3.73GNDD341 pKa = 3.28EE342 pKa = 4.64FIIADD347 pKa = 3.75LDD349 pKa = 4.41EE350 pKa = 4.28IHH352 pKa = 7.09LEE354 pKa = 4.04QEE356 pKa = 4.31EE357 pKa = 4.38LQGGAGTDD365 pKa = 2.91RR366 pKa = 11.84LTLQEE371 pKa = 4.44SGAVDD376 pKa = 4.07LSKK379 pKa = 11.41VLMFDD384 pKa = 3.4MEE386 pKa = 4.42EE387 pKa = 4.73LEE389 pKa = 4.79LSNGGNTVTMVRR401 pKa = 11.84EE402 pKa = 3.92QLLQLTNIVPAAQGDD417 pKa = 4.64SILKK421 pKa = 9.47LAQAATLDD429 pKa = 4.14LEE431 pKa = 5.17QMPQLQAFNKK441 pKa = 9.91IYY443 pKa = 10.68GATTGSNDD451 pKa = 3.38LLGYY455 pKa = 10.78AGDD458 pKa = 4.7DD459 pKa = 3.57YY460 pKa = 11.88LVGGKK465 pKa = 9.44NEE467 pKa = 3.9DD468 pKa = 3.44QLRR471 pKa = 11.84GGEE474 pKa = 4.18GVDD477 pKa = 3.52YY478 pKa = 11.56LEE480 pKa = 5.03GGEE483 pKa = 4.27GADD486 pKa = 2.46IYY488 pKa = 11.47VFNRR492 pKa = 11.84ADD494 pKa = 3.45YY495 pKa = 9.37SQQSVGATDD504 pKa = 4.05FVGDD508 pKa = 4.66TIHH511 pKa = 7.17GLDD514 pKa = 3.74FTQGDD519 pKa = 4.56KK520 pKa = 10.71IRR522 pKa = 11.84LEE524 pKa = 4.34APLSGSTQALSLEE537 pKa = 4.26SGFYY541 pKa = 10.61SSLTEE546 pKa = 4.38TIQANATIQSAFAGDD561 pKa = 4.15DD562 pKa = 3.17VDD564 pKa = 5.73AVLLQVKK571 pKa = 10.02AGTAAGYY578 pKa = 9.36YY579 pKa = 10.54LLVEE583 pKa = 5.08EE584 pKa = 5.11YY585 pKa = 10.81NADD588 pKa = 3.84YY589 pKa = 11.06QPADD593 pKa = 3.62ATDD596 pKa = 3.72TTDD599 pKa = 4.36DD600 pKa = 3.81AASLVNTEE608 pKa = 4.28TDD610 pKa = 3.28TLNYY614 pKa = 10.6DD615 pKa = 3.64PASDD619 pKa = 4.95LIIRR623 pKa = 11.84LTGIEE628 pKa = 5.01SLNQLNGFSTSVFII642 pKa = 5.76
Molecular weight: 69.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H6U170|A0A1H6U170_9GAMM Acetyl-CoA C-acetyltransferase OS=Pseudospirillum japonicum OX=64971 GN=SAMN05421831_11252 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 9.97RR12 pKa = 11.84KK13 pKa = 8.91RR14 pKa = 11.84VHH16 pKa = 6.29GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 9.97RR12 pKa = 11.84KK13 pKa = 8.91RR14 pKa = 11.84VHH16 pKa = 6.29GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
820087 |
31 |
1776 |
343.9 |
38.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.38 ± 0.055 | 1.029 ± 0.017 |
4.964 ± 0.036 | 5.909 ± 0.049 |
3.491 ± 0.03 | 6.437 ± 0.05 |
2.777 ± 0.029 | 5.503 ± 0.035 |
3.881 ± 0.042 | 11.777 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.273 ± 0.022 | 3.086 ± 0.03 |
4.423 ± 0.037 | 7.071 ± 0.069 |
5.289 ± 0.038 | 5.526 ± 0.04 |
5.333 ± 0.044 | 6.397 ± 0.039 |
1.443 ± 0.023 | 3.011 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
