
Vulpes vulpes papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Treisetapapillomavirus; Treisetapapillomavirus 1
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
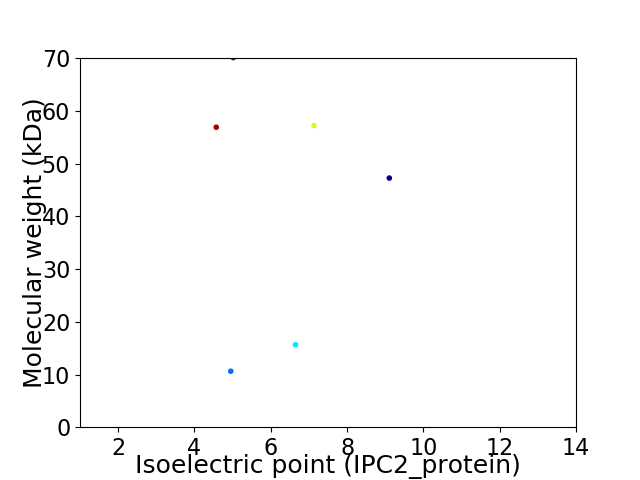
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7BW90|A0A0A7BW90_9PAPI Replication protein E1 OS=Vulpes vulpes papillomavirus 1 OX=1163709 GN=E1 PE=3 SV=1
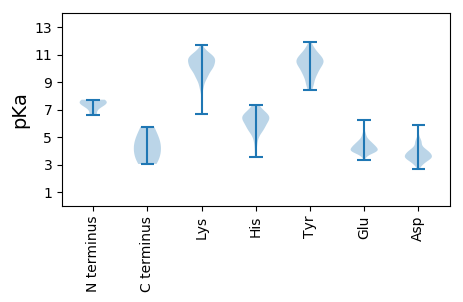
MM1 pKa = 7.68LPRR4 pKa = 11.84KK5 pKa = 9.11RR6 pKa = 11.84AKK8 pKa = 10.22RR9 pKa = 11.84DD10 pKa = 3.49TVDD13 pKa = 3.08NLYY16 pKa = 10.09RR17 pKa = 11.84QCQITGNCPPDD28 pKa = 3.46VKK30 pKa = 11.11NKK32 pKa = 10.12VEE34 pKa = 4.18GTTLADD40 pKa = 3.12ILSKK44 pKa = 10.38IFSSIIYY51 pKa = 9.31TGGLGIGTGKK61 pKa = 8.8GTGGTGGYY69 pKa = 9.4RR70 pKa = 11.84PLGGTTGSRR79 pKa = 11.84GPGTVSRR86 pKa = 11.84PNIPVDD92 pKa = 4.07PIGPADD98 pKa = 3.96ILPINPVEE106 pKa = 4.31PNASSIVPLNEE117 pKa = 3.96GDD119 pKa = 3.41PGIIIEE125 pKa = 4.79PPGTVNGADD134 pKa = 3.38VEE136 pKa = 4.48LSVVTSIDD144 pKa = 3.47PVSDD148 pKa = 3.41VSNASIPAPTGAGSDD163 pKa = 3.38GDD165 pKa = 3.9AAIIDD170 pKa = 4.18VQTSTRR176 pKa = 11.84TTNAVRR182 pKa = 11.84STQRR186 pKa = 11.84FNNPVFEE193 pKa = 4.48TVAIVHH199 pKa = 6.5RR200 pKa = 11.84SFGEE204 pKa = 3.83SSEE207 pKa = 3.99NLNVVVDD214 pKa = 3.98PTILQDD220 pKa = 3.54TVIGGDD226 pKa = 2.9EE227 pKa = 4.45GEE229 pKa = 4.65FIEE232 pKa = 5.87LDD234 pKa = 3.05ILGRR238 pKa = 11.84PSQFEE243 pKa = 3.99IVEE246 pKa = 4.21GEE248 pKa = 4.31GPKK251 pKa = 10.0TSTPIEE257 pKa = 4.1RR258 pKa = 11.84LAGPFKK264 pKa = 10.65RR265 pKa = 11.84ARR267 pKa = 11.84EE268 pKa = 4.02LYY270 pKa = 9.81NRR272 pKa = 11.84RR273 pKa = 11.84IKK275 pKa = 10.19QIPTQNPLFLGRR287 pKa = 11.84AAEE290 pKa = 3.88AVEE293 pKa = 5.0FGFEE297 pKa = 3.82NPAYY301 pKa = 9.66EE302 pKa = 5.23DD303 pKa = 4.49DD304 pKa = 3.79VTLTFEE310 pKa = 4.96RR311 pKa = 11.84DD312 pKa = 3.37LDD314 pKa = 3.89EE315 pKa = 5.1LAAAPVAAPDD325 pKa = 4.17PDD327 pKa = 3.58FADD330 pKa = 3.07IVKK333 pKa = 10.59LSRR336 pKa = 11.84PRR338 pKa = 11.84LSEE341 pKa = 3.97PEE343 pKa = 4.02PGRR346 pKa = 11.84VRR348 pKa = 11.84YY349 pKa = 9.75SRR351 pKa = 11.84LGRR354 pKa = 11.84RR355 pKa = 11.84GTMTLRR361 pKa = 11.84SGVQIGEE368 pKa = 4.05EE369 pKa = 3.92VHH371 pKa = 7.03FYY373 pKa = 11.04RR374 pKa = 11.84DD375 pKa = 3.26LSTIDD380 pKa = 3.55DD381 pKa = 3.9AQGIEE386 pKa = 4.3LSVLGHH392 pKa = 5.33QSGEE396 pKa = 4.07EE397 pKa = 4.1VIIDD401 pKa = 3.99PLSEE405 pKa = 4.12STVIDD410 pKa = 3.66AEE412 pKa = 4.58NISEE416 pKa = 4.14PQFDD420 pKa = 3.86YY421 pKa = 11.5DD422 pKa = 3.86EE423 pKa = 4.91YY424 pKa = 11.46EE425 pKa = 4.14HH426 pKa = 7.12LLLDD430 pKa = 4.13EE431 pKa = 4.78LSEE434 pKa = 4.61DD435 pKa = 3.91FSNSHH440 pKa = 6.54LVLNSTGRR448 pKa = 11.84NSSLTMPSMPPGTPLKK464 pKa = 10.61IFIDD468 pKa = 4.46DD469 pKa = 3.82YY470 pKa = 11.47GSLIVAHH477 pKa = 6.51SSSTEE482 pKa = 3.73SDD484 pKa = 4.7DD485 pKa = 3.62IPSTIWEE492 pKa = 4.23NLQPVIVVSPFEE504 pKa = 4.07SFDD507 pKa = 4.0YY508 pKa = 10.75NLHH511 pKa = 6.08PSLRR515 pKa = 11.84RR516 pKa = 11.84KK517 pKa = 8.88KK518 pKa = 10.06RR519 pKa = 11.84KK520 pKa = 9.24RR521 pKa = 11.84PYY523 pKa = 10.36FFF525 pKa = 5.75
MM1 pKa = 7.68LPRR4 pKa = 11.84KK5 pKa = 9.11RR6 pKa = 11.84AKK8 pKa = 10.22RR9 pKa = 11.84DD10 pKa = 3.49TVDD13 pKa = 3.08NLYY16 pKa = 10.09RR17 pKa = 11.84QCQITGNCPPDD28 pKa = 3.46VKK30 pKa = 11.11NKK32 pKa = 10.12VEE34 pKa = 4.18GTTLADD40 pKa = 3.12ILSKK44 pKa = 10.38IFSSIIYY51 pKa = 9.31TGGLGIGTGKK61 pKa = 8.8GTGGTGGYY69 pKa = 9.4RR70 pKa = 11.84PLGGTTGSRR79 pKa = 11.84GPGTVSRR86 pKa = 11.84PNIPVDD92 pKa = 4.07PIGPADD98 pKa = 3.96ILPINPVEE106 pKa = 4.31PNASSIVPLNEE117 pKa = 3.96GDD119 pKa = 3.41PGIIIEE125 pKa = 4.79PPGTVNGADD134 pKa = 3.38VEE136 pKa = 4.48LSVVTSIDD144 pKa = 3.47PVSDD148 pKa = 3.41VSNASIPAPTGAGSDD163 pKa = 3.38GDD165 pKa = 3.9AAIIDD170 pKa = 4.18VQTSTRR176 pKa = 11.84TTNAVRR182 pKa = 11.84STQRR186 pKa = 11.84FNNPVFEE193 pKa = 4.48TVAIVHH199 pKa = 6.5RR200 pKa = 11.84SFGEE204 pKa = 3.83SSEE207 pKa = 3.99NLNVVVDD214 pKa = 3.98PTILQDD220 pKa = 3.54TVIGGDD226 pKa = 2.9EE227 pKa = 4.45GEE229 pKa = 4.65FIEE232 pKa = 5.87LDD234 pKa = 3.05ILGRR238 pKa = 11.84PSQFEE243 pKa = 3.99IVEE246 pKa = 4.21GEE248 pKa = 4.31GPKK251 pKa = 10.0TSTPIEE257 pKa = 4.1RR258 pKa = 11.84LAGPFKK264 pKa = 10.65RR265 pKa = 11.84ARR267 pKa = 11.84EE268 pKa = 4.02LYY270 pKa = 9.81NRR272 pKa = 11.84RR273 pKa = 11.84IKK275 pKa = 10.19QIPTQNPLFLGRR287 pKa = 11.84AAEE290 pKa = 3.88AVEE293 pKa = 5.0FGFEE297 pKa = 3.82NPAYY301 pKa = 9.66EE302 pKa = 5.23DD303 pKa = 4.49DD304 pKa = 3.79VTLTFEE310 pKa = 4.96RR311 pKa = 11.84DD312 pKa = 3.37LDD314 pKa = 3.89EE315 pKa = 5.1LAAAPVAAPDD325 pKa = 4.17PDD327 pKa = 3.58FADD330 pKa = 3.07IVKK333 pKa = 10.59LSRR336 pKa = 11.84PRR338 pKa = 11.84LSEE341 pKa = 3.97PEE343 pKa = 4.02PGRR346 pKa = 11.84VRR348 pKa = 11.84YY349 pKa = 9.75SRR351 pKa = 11.84LGRR354 pKa = 11.84RR355 pKa = 11.84GTMTLRR361 pKa = 11.84SGVQIGEE368 pKa = 4.05EE369 pKa = 3.92VHH371 pKa = 7.03FYY373 pKa = 11.04RR374 pKa = 11.84DD375 pKa = 3.26LSTIDD380 pKa = 3.55DD381 pKa = 3.9AQGIEE386 pKa = 4.3LSVLGHH392 pKa = 5.33QSGEE396 pKa = 4.07EE397 pKa = 4.1VIIDD401 pKa = 3.99PLSEE405 pKa = 4.12STVIDD410 pKa = 3.66AEE412 pKa = 4.58NISEE416 pKa = 4.14PQFDD420 pKa = 3.86YY421 pKa = 11.5DD422 pKa = 3.86EE423 pKa = 4.91YY424 pKa = 11.46EE425 pKa = 4.14HH426 pKa = 7.12LLLDD430 pKa = 4.13EE431 pKa = 4.78LSEE434 pKa = 4.61DD435 pKa = 3.91FSNSHH440 pKa = 6.54LVLNSTGRR448 pKa = 11.84NSSLTMPSMPPGTPLKK464 pKa = 10.61IFIDD468 pKa = 4.46DD469 pKa = 3.82YY470 pKa = 11.47GSLIVAHH477 pKa = 6.51SSSTEE482 pKa = 3.73SDD484 pKa = 4.7DD485 pKa = 3.62IPSTIWEE492 pKa = 4.23NLQPVIVVSPFEE504 pKa = 4.07SFDD507 pKa = 4.0YY508 pKa = 10.75NLHH511 pKa = 6.08PSLRR515 pKa = 11.84RR516 pKa = 11.84KK517 pKa = 8.88KK518 pKa = 10.06RR519 pKa = 11.84KK520 pKa = 9.24RR521 pKa = 11.84PYY523 pKa = 10.36FFF525 pKa = 5.75
Molecular weight: 56.94 kDa
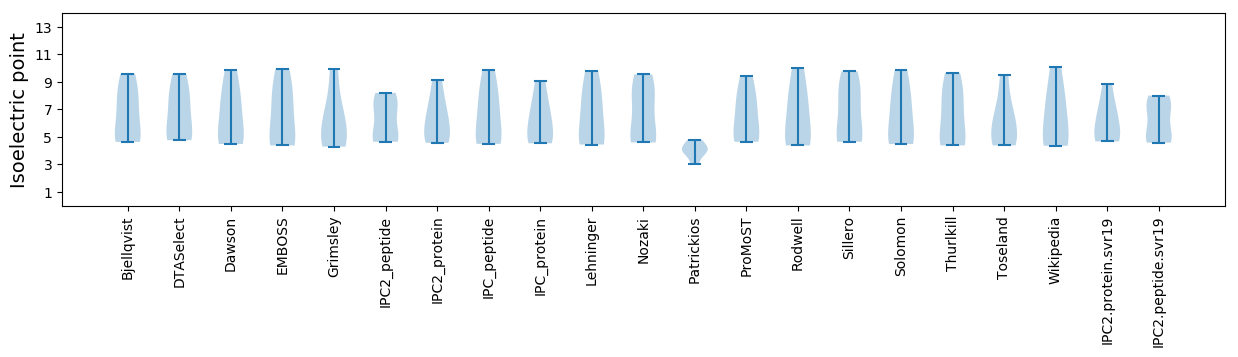
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7BWG9|A0A0A7BWG9_9PAPI Regulatory protein E2 OS=Vulpes vulpes papillomavirus 1 OX=1163709 GN=E2 PE=3 SV=1
MM1 pKa = 6.59EE2 pKa = 4.56TLRR5 pKa = 11.84EE6 pKa = 3.97RR7 pKa = 11.84FDD9 pKa = 3.9ALQEE13 pKa = 4.04QLLTLYY19 pKa = 10.08EE20 pKa = 4.08QEE22 pKa = 4.45NNDD25 pKa = 4.37LEE27 pKa = 4.55SQLLHH32 pKa = 6.44WKK34 pKa = 9.31LQRR37 pKa = 11.84KK38 pKa = 8.55EE39 pKa = 4.06SALMYY44 pKa = 9.03YY45 pKa = 10.1ARR47 pKa = 11.84KK48 pKa = 10.0NGLASLGMQRR58 pKa = 11.84LPSLQVSEE66 pKa = 4.43QQAKK70 pKa = 8.76QAILMTLYY78 pKa = 10.9LEE80 pKa = 4.79SLSKK84 pKa = 9.62TPYY87 pKa = 10.61AKK89 pKa = 10.35EE90 pKa = 3.4HH91 pKa = 5.88WTLQDD96 pKa = 2.91TSYY99 pKa = 10.92EE100 pKa = 4.08RR101 pKa = 11.84FMGQPPYY108 pKa = 10.38CFKK111 pKa = 10.87KK112 pKa = 10.75GPFTVDD118 pKa = 2.65VVYY121 pKa = 11.04DD122 pKa = 3.65KK123 pKa = 11.34DD124 pKa = 4.09SNNMFSYY131 pKa = 10.54TNYY134 pKa = 10.53NFIYY138 pKa = 10.48YY139 pKa = 9.66QDD141 pKa = 4.09EE142 pKa = 4.32DD143 pKa = 4.57EE144 pKa = 4.69NWHH147 pKa = 5.59KK148 pKa = 9.81TSGEE152 pKa = 3.83VSYY155 pKa = 11.18EE156 pKa = 3.48GLYY159 pKa = 8.53YY160 pKa = 10.21TDD162 pKa = 3.29YY163 pKa = 10.75KK164 pKa = 10.57GRR166 pKa = 11.84SVYY169 pKa = 10.0FVYY172 pKa = 10.64FEE174 pKa = 5.24KK175 pKa = 10.78DD176 pKa = 2.97AQLYY180 pKa = 8.39SQYY183 pKa = 9.82GTWEE187 pKa = 3.9VHH189 pKa = 6.57FKK191 pKa = 10.72NQTISASVVSTSTGTGSGAYY211 pKa = 9.26DD212 pKa = 4.1SPDD215 pKa = 3.02SSPPGRR221 pKa = 11.84RR222 pKa = 11.84STGRR226 pKa = 11.84PRR228 pKa = 11.84VSEE231 pKa = 3.84ARR233 pKa = 11.84QPANTSPTAYY243 pKa = 9.64PEE245 pKa = 4.36ASEE248 pKa = 4.0KK249 pKa = 9.53TALRR253 pKa = 11.84KK254 pKa = 9.84RR255 pKa = 11.84PGPGPGQTSPSPSAGAPKK273 pKa = 9.91RR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84GEE279 pKa = 3.81RR280 pKa = 11.84EE281 pKa = 3.55QVPRR285 pKa = 11.84QLRR288 pKa = 11.84STPSVSSGTPWPSPGQVGRR307 pKa = 11.84RR308 pKa = 11.84SRR310 pKa = 11.84LPPRR314 pKa = 11.84HH315 pKa = 5.56NSSRR319 pKa = 11.84LRR321 pKa = 11.84RR322 pKa = 11.84LQAEE326 pKa = 3.99AWDD329 pKa = 4.1PPVILLKK336 pKa = 11.05GKK338 pKa = 10.35ANNLKK343 pKa = 9.99CWRR346 pKa = 11.84NRR348 pKa = 11.84VKK350 pKa = 10.51QRR352 pKa = 11.84NLDD355 pKa = 3.67FKK357 pKa = 10.95IVFSSLFNWIGLSHH371 pKa = 6.55TEE373 pKa = 3.98HH374 pKa = 6.64GKK376 pKa = 9.56HH377 pKa = 6.23RR378 pKa = 11.84MLLAFDD384 pKa = 4.21GEE386 pKa = 4.33KK387 pKa = 10.39EE388 pKa = 3.8RR389 pKa = 11.84RR390 pKa = 11.84LFLDD394 pKa = 3.48TVKK397 pKa = 10.63LPKK400 pKa = 10.51GVTYY404 pKa = 10.95SLGNLDD410 pKa = 3.96SLL412 pKa = 4.83
MM1 pKa = 6.59EE2 pKa = 4.56TLRR5 pKa = 11.84EE6 pKa = 3.97RR7 pKa = 11.84FDD9 pKa = 3.9ALQEE13 pKa = 4.04QLLTLYY19 pKa = 10.08EE20 pKa = 4.08QEE22 pKa = 4.45NNDD25 pKa = 4.37LEE27 pKa = 4.55SQLLHH32 pKa = 6.44WKK34 pKa = 9.31LQRR37 pKa = 11.84KK38 pKa = 8.55EE39 pKa = 4.06SALMYY44 pKa = 9.03YY45 pKa = 10.1ARR47 pKa = 11.84KK48 pKa = 10.0NGLASLGMQRR58 pKa = 11.84LPSLQVSEE66 pKa = 4.43QQAKK70 pKa = 8.76QAILMTLYY78 pKa = 10.9LEE80 pKa = 4.79SLSKK84 pKa = 9.62TPYY87 pKa = 10.61AKK89 pKa = 10.35EE90 pKa = 3.4HH91 pKa = 5.88WTLQDD96 pKa = 2.91TSYY99 pKa = 10.92EE100 pKa = 4.08RR101 pKa = 11.84FMGQPPYY108 pKa = 10.38CFKK111 pKa = 10.87KK112 pKa = 10.75GPFTVDD118 pKa = 2.65VVYY121 pKa = 11.04DD122 pKa = 3.65KK123 pKa = 11.34DD124 pKa = 4.09SNNMFSYY131 pKa = 10.54TNYY134 pKa = 10.53NFIYY138 pKa = 10.48YY139 pKa = 9.66QDD141 pKa = 4.09EE142 pKa = 4.32DD143 pKa = 4.57EE144 pKa = 4.69NWHH147 pKa = 5.59KK148 pKa = 9.81TSGEE152 pKa = 3.83VSYY155 pKa = 11.18EE156 pKa = 3.48GLYY159 pKa = 8.53YY160 pKa = 10.21TDD162 pKa = 3.29YY163 pKa = 10.75KK164 pKa = 10.57GRR166 pKa = 11.84SVYY169 pKa = 10.0FVYY172 pKa = 10.64FEE174 pKa = 5.24KK175 pKa = 10.78DD176 pKa = 2.97AQLYY180 pKa = 8.39SQYY183 pKa = 9.82GTWEE187 pKa = 3.9VHH189 pKa = 6.57FKK191 pKa = 10.72NQTISASVVSTSTGTGSGAYY211 pKa = 9.26DD212 pKa = 4.1SPDD215 pKa = 3.02SSPPGRR221 pKa = 11.84RR222 pKa = 11.84STGRR226 pKa = 11.84PRR228 pKa = 11.84VSEE231 pKa = 3.84ARR233 pKa = 11.84QPANTSPTAYY243 pKa = 9.64PEE245 pKa = 4.36ASEE248 pKa = 4.0KK249 pKa = 9.53TALRR253 pKa = 11.84KK254 pKa = 9.84RR255 pKa = 11.84PGPGPGQTSPSPSAGAPKK273 pKa = 9.91RR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84GEE279 pKa = 3.81RR280 pKa = 11.84EE281 pKa = 3.55QVPRR285 pKa = 11.84QLRR288 pKa = 11.84STPSVSSGTPWPSPGQVGRR307 pKa = 11.84RR308 pKa = 11.84SRR310 pKa = 11.84LPPRR314 pKa = 11.84HH315 pKa = 5.56NSSRR319 pKa = 11.84LRR321 pKa = 11.84RR322 pKa = 11.84LQAEE326 pKa = 3.99AWDD329 pKa = 4.1PPVILLKK336 pKa = 11.05GKK338 pKa = 10.35ANNLKK343 pKa = 9.99CWRR346 pKa = 11.84NRR348 pKa = 11.84VKK350 pKa = 10.51QRR352 pKa = 11.84NLDD355 pKa = 3.67FKK357 pKa = 10.95IVFSSLFNWIGLSHH371 pKa = 6.55TEE373 pKa = 3.98HH374 pKa = 6.64GKK376 pKa = 9.56HH377 pKa = 6.23RR378 pKa = 11.84MLLAFDD384 pKa = 4.21GEE386 pKa = 4.33KK387 pKa = 10.39EE388 pKa = 3.8RR389 pKa = 11.84RR390 pKa = 11.84LFLDD394 pKa = 3.48TVKK397 pKa = 10.63LPKK400 pKa = 10.51GVTYY404 pKa = 10.95SLGNLDD410 pKa = 3.96SLL412 pKa = 4.83
Molecular weight: 47.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2303 |
96 |
623 |
383.8 |
42.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.037 ± 0.387 | 2.084 ± 0.739 |
6.036 ± 0.548 | 6.34 ± 0.335 |
4.516 ± 0.354 | 6.687 ± 0.684 |
1.693 ± 0.237 | 4.342 ± 0.864 |
5.211 ± 0.762 | 9.857 ± 0.835 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.563 ± 0.295 | 4.907 ± 0.562 |
6.426 ± 0.743 | 3.995 ± 0.503 |
5.775 ± 0.844 | 8.641 ± 0.61 |
6.296 ± 0.381 | 6.166 ± 0.45 |
1.259 ± 0.308 | 3.17 ± 0.536 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
