
Proteome-pI 2.0 - Proteome Isoelectric Point Database is a database of pre-computed isoelectric points for proteomes from different model organisms (20, 115 species).
A full list of organisms can be seen here
General statistics for analyzed proteomes
|
|
Number of proteomes |
Total number of proteins |
Mean number of proteins ± SD |
Mean size of proteins ± SD |
Mean mw of proteins ± SD |
|
Viruses Archaea Bacteria Eukaryote (all isoforms) Eukaryote (main isoform) Eukaryote (minor isoforms) |
10,064 331 8,108 1,612 1,612 637 |
518,140 767,951 30,290,647 29,752,296 25,437,198 4,315,098 |
51 ± 85 2,320 ± 1,263 3,736 ± 1,785 18,457 ± 16,804 15,780 ± 11,138 6,774 ± 14,244 |
237 ± 300 278 ± 211 320 ± 246 467 ± 471 438 ± 420 638 ± 676 |
26.6 ± 33.2 30.6 ± 23.1 35.1 ± 26.8 52.1 ± 52.4 48.8 ± 46.7 71.2 ± 75.4 |
|
|
IDM number of proteins |
Median number of protein |
IDM size of proteins |
Median size of protein |
IDM mw of proteins |
Median mw of protein |
|
Viruses Archaea Bacteria Eukaryote (all isoforms) Eukaryote (main isoform) Eukaryote (minor isoforms) |
35 2,265 3,583 15,677 14,121 3,916 |
17 2,164 3,503 13,001 12,791 1,000 |
182 249 287 391 370 534 |
145 234 274 353 336 476 |
20.4 27.4 31.4 43.6 41.2 59.7 |
16.4 25.8 30.0 39.4 37.5 53.2 |
As one can see Viruses have the smallest proteomes (coding usually only a handful number of proteins) with compacted proteins. Then Archaea step in with relatively small proteomes (~2.3k) and short proteins (234-278 aa). The next group, Bacteria, code bigger proteomes (~3.7k) and longer proteins (274-320 aa).
Eukaryotes on the other hand are the most sophisticated, having the biggest proteomes (~18k) and the longest proteins (336-476 aa). Moreover, many of them possess multiple splicing isoforms which in some cases can significantly increase proteome complexity (e.g. in humans 20,600 proteins vs. 79,500 additional isoforms)
General isoelectric point statistics
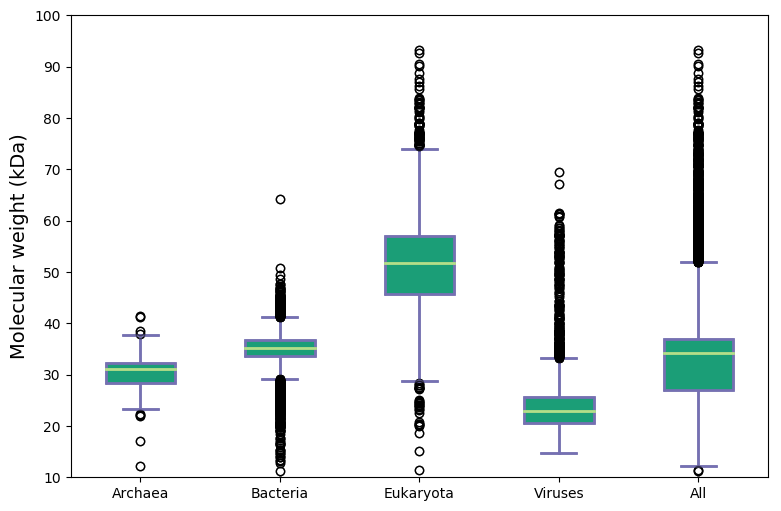
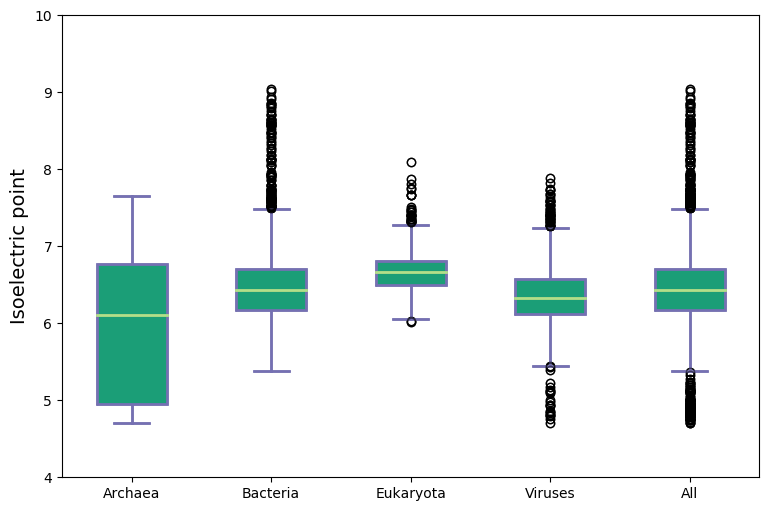
Molecular weight and isoelectric points across kingdoms
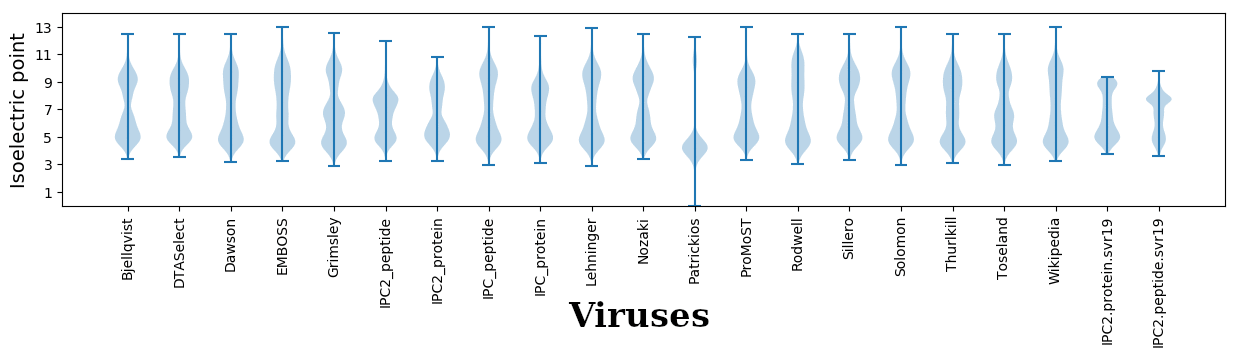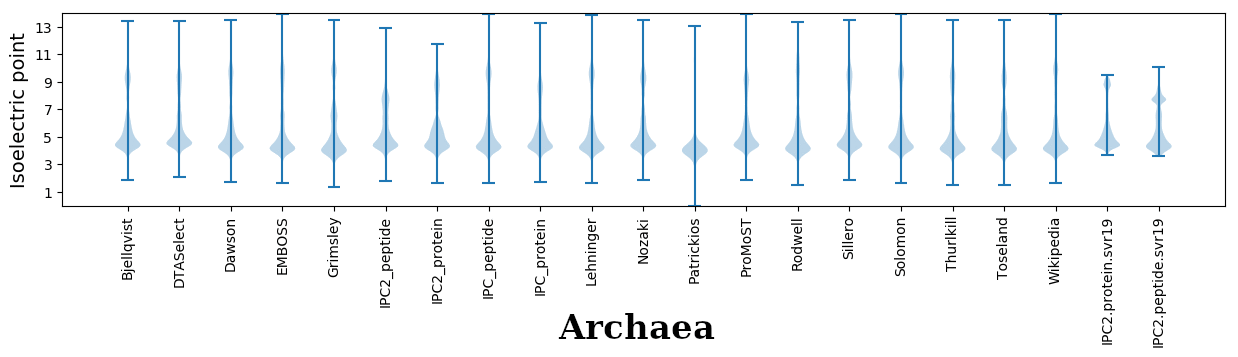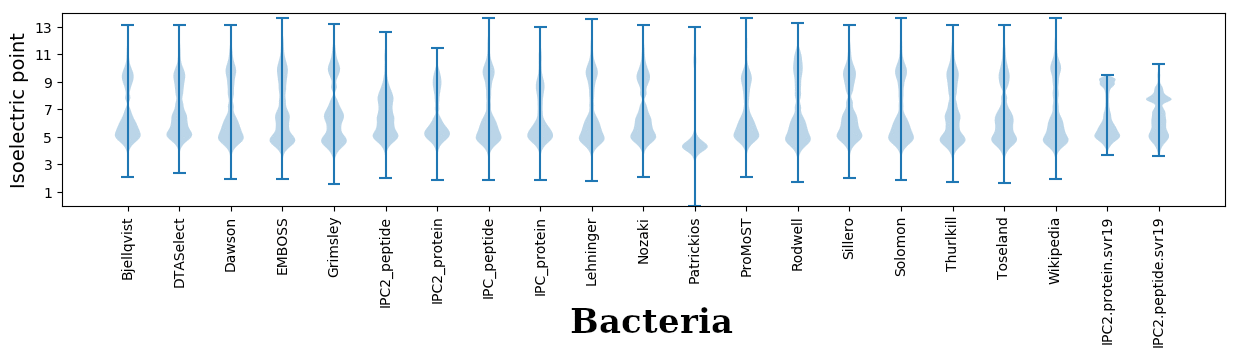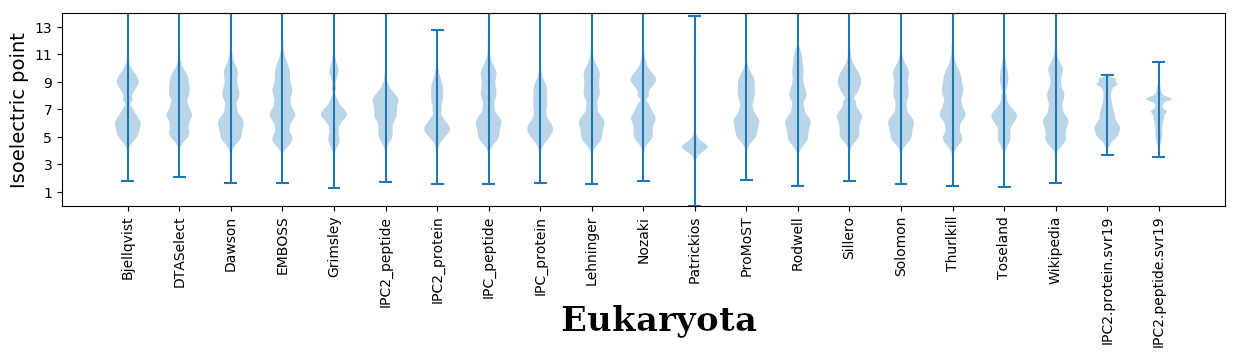
Isoelectric points for different methods
Dissociation contant (pKa) predictions according to charge location
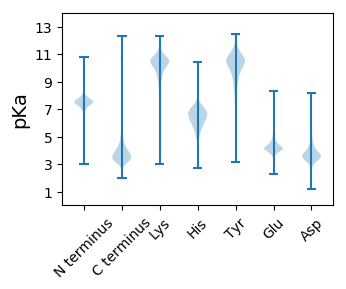
Total pKa predictions processed: 39534350 --- N-terminal Statistics (with 95% Bootstrap CI) --- AA | Count | Mean pKa | 95% CI (Bootstrap) ------------------------------------------------------ A | 90817 | 7.6940 | [7.6863, 7.7012] C | 2634 | 7.4084 | [7.3945, 7.4225] F | 3880 | 7.2807 | [7.2694, 7.2925] G | 78233 | 7.2744 | [7.2624, 7.2865] I | 7053 | 7.3942 | [7.3833, 7.4069] L | 6214 | 7.2573 | [7.2433, 7.2705] M | 296068 | 7.4535 | [7.4473, 7.4600] N | 5260 | 7.2081 | [7.1944, 7.2225] P | 15874 | 7.3866 | [7.3791, 7.3942] Q | 13562 | 7.0611 | [7.0473, 7.0751] R | 6931 | 7.5516 | [7.5395, 7.5632] S | 42343 | 6.5932 | [6.5822, 6.6060] T | 13451 | 6.8833 | [6.8717, 6.8943] V | 9015 | 6.8125 | [6.8005, 6.8243] W | 691 | 7.2039 | [7.1574, 7.2508] X | 9143 | 6.9416 | [6.9254, 6.9553] --- Middle Statistics (with 95% Bootstrap CI) --- AA | Count | Mean pKa | 95% CI (Bootstrap) ------------------------------------------------------ D | 8798307 | 3.7408 | [3.7286, 3.7530] E | 10409080 | 4.2426 | [4.2345, 4.2506] H | 4125923 | 6.3540 | [6.3384, 6.3677] K | 9578878 | 10.1301 | [10.1150, 10.1466] Y | 5419824 | 10.0584 | [10.0363, 10.0777] --- C-terminal Statistics (with 95% Bootstrap CI) --- AA | Count | Mean pKa | 95% CI (Bootstrap) ------------------------------------------------------ A | 252699 | 4.1999 | [4.1838, 4.2156] C | 10163 | 3.9616 | [3.9435, 3.9779] F | 20887 | 4.0889 | [4.0752, 4.1038] G | 34657 | 3.5490 | [3.5408, 3.5571] I | 17640 | 4.2118 | [4.1990, 4.2256] L | 50384 | 3.8461 | [3.8350, 3.8572] M | 9651 | 4.8977 | [4.8740, 4.9233] N | 23372 | 3.5951 | [3.5869, 3.6040] P | 18875 | 3.8796 | [3.8679, 3.8918] Q | 27009 | 3.5865 | [3.5746, 3.5985] R | 39070 | 3.9662 | [3.9536, 3.9795] S | 41790 | 3.3903 | [3.3831, 3.3969] T | 15787 | 3.8216 | [3.8107, 3.8316] V | 23929 | 3.3292 | [3.3210, 3.3373] W | 7213 | 3.6729 | [3.6545, 3.6910] X | 8043 | 4.9259 | [4.9017, 4.9490]
Amino acid frequency across kingdoms
| Kingdom | Ala | Cys | Asp | Glu | Phe | Gly | His | Ile | Lys | Leu | Met | Asn | Pro | Gln | Arg | Ser | Thr | Val | Trp | Tyr | Xaa | Total |
TXT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Viruses | 7.81Ala: 7.806 ± 0.078 |
1.29Cys: 1.287 ± 0.018 |
6.20Asp: 6.204 ± 0.016 |
6.46Glu: 6.457 ± 0.031 |
3.91Phe: 3.910 ± 0.019 |
6.72Gly: 6.721 ± 0.030 |
1.96His: 1.962 ± 0.011 |
6.05Ile: 6.048 ± 0.050 |
6.24Lys: 6.237 ± 0.051 |
8.28Leu: 8.281 ± 0.014 |
2.51Met: 2.511 ± 0.008 |
4.99Asn: 4.990 ± 0.050 |
4.25Pro: 4.247 ± 0.028 |
3.62Gln: 3.620 ± 0.014 |
5.31Arg: 5.306 ± 0.042 |
6.47Ser: 6.474 ± 0.017 |
6.14Thr: 6.141 ± 0.015 |
6.66Val: 6.664 ± 0.021 |
1.42Trp: 1.416 ± 0.009 |
3.71Tyr: 3.714 ± 0.004 |
0.004Xaa: 0.004 ± 0.001 |
122,870,810 |
|
| Archaea | 8.95Ala: 8.951 ± 0.136 |
0.90Cys: 0.899 ± 0.020 |
7.00Asp: 6.995 ± 0.100 |
6.46Glu: 6.457 ± 0.031 |
3.65Phe: 3.650 ± 0.031 |
7.84Gly: 7.841 ± 0.060 |
1.86His: 1.860 ± 0.018 |
6.03Ile: 6.030 ± 0.135 |
4.18Lys: 4.184 ± 0.156 |
9.11Leu: 9.107 ± 0.036 |
2.14Met: 2.138 ± 0.030 |
3.36Asn: 3.358 ± 0.083 |
4.36Pro: 4.363 ± 0.029 |
2.48Gln: 2.477 ± 0.033 |
5.83Arg: 5.833 ± 0.071 |
6.12Ser: 6.118 ± 0.059 |
5.84Thr: 5.842 ± 0.048 |
8.16Val: 8.164 ± 0.071 |
1.06Trp: 1.058 ± 0.009 |
3.18Tyr: 3.183 ± 0.037 |
0.010Xaa: 0.010 ± 0.004 |
213,285,886 |
|
| Bacteria | 10.64Ala: 10.638 ± 0.034 |
0.90Cys: 0.902 ± 0.003 |
5.67Asp: 5.668 ± 0.006 |
6.06Glu: 6.056 ± 0.009 |
3.76Phe: 3.764 ± 0.009 |
8.01Gly: 8.008 ± 0.014 |
2.08His: 2.080 ± 0.003 |
5.52Ile: 5.524 ± 0.022 |
4.23Lys: 4.225 ± 0.025 |
10.12Leu: 10.117 ± 0.008 |
2.31Met: 2.312 ± 0.005 |
3.35Asn: 3.353 ± 0.017 |
4.82Pro: 4.815 ± 0.012 |
3.49Gln: 3.491 ± 0.010 |
6.18Arg: 6.177 ± 0.020 |
5.75Ser: 5.746 ± 0.008 |
5.58Thr: 5.578 ± 0.006 |
7.42Val: 7.422 ± 0.012 |
1.31Trp: 1.313 ± 0.003 |
2.81Tyr: 2.809 ± 0.010 |
0.001Xaa: 0.001 ± 0.000 |
9,693,905,784 |
|
| Eukaryota | 7.38Ala: 7.376 ± 0.045 |
1.85Cys: 1.852 ± 0.012 |
5.34Asp: 5.341 ± 0.011 |
6.55Glu: 6.555 ± 0.017 |
3.79Phe: 3.794 ± 0.012 |
6.35Gly: 6.354 ± 0.021 |
2.50His: 2.498 ± 0.006 |
4.94Ile: 4.942 ± 0.024 |
5.64Lys: 5.640 ± 0.024 |
9.39Leu: 9.385 ± 0.015 |
2.27Met: 2.268 ± 0.006 |
4.13Asn: 4.134 ± 0.027 |
5.56Pro: 5.561 ± 0.022 |
4.27Gln: 4.274 ± 0.023 |
5.71Arg: 5.705 ± 0.018 |
8.45Ser: 8.454 ± 0.015 |
5.56Thr: 5.556 ± 0.016 |
6.24Val: 6.238 ± 0.013 |
1.24Trp: 1.239 ± 0.005 |
2.81Tyr: 2.806 ± 0.010 |
0.028Xaa: 0.028 ± 0.003 |
13,901,635,566 |
|
| All | 8.72Ala: 8.715 ± 0.045 |
1.46Cys: 1.455 ± 0.014 |
5.49Asp: 5.493 ± 0.008 |
6.36Glu: 6.364 ± 0.012 |
3.78Phe: 3.781 ± 0.008 |
7.04Gly: 7.039 ± 0.019 |
2.32His: 2.321 ± 0.006 |
5.19Ile: 5.193 ± 0.018 |
5.06Lys: 5.055 ± 0.022 |
9.67Leu: 9.674 ± 0.009 |
2.29Met: 2.286 ± 0.004 |
3.82Asn: 3.815 ± 0.017 |
5.24Pro: 5.241 ± 0.015 |
3.94Gln: 3.938 ± 0.016 |
5.90Arg: 5.896 ± 0.014 |
7.33Ser: 7.326 ± 0.026 |
5.57Thr: 5.571 ± 0.010 |
6.74Val: 6.738 ± 0.013 |
1.27Trp: 1.268 ± 0.003 |
2.82Tyr: 2.815 ± 0.007 |
0.017Xaa: 0.017 ± 0.002 |
23,931,698,046 |
More amino acid statistics
Dipeptides |
|||||
|---|---|---|---|---|---|
Tripeptides |
Proteome-pI 2.0 is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
