
Tortoise microvirus 55
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
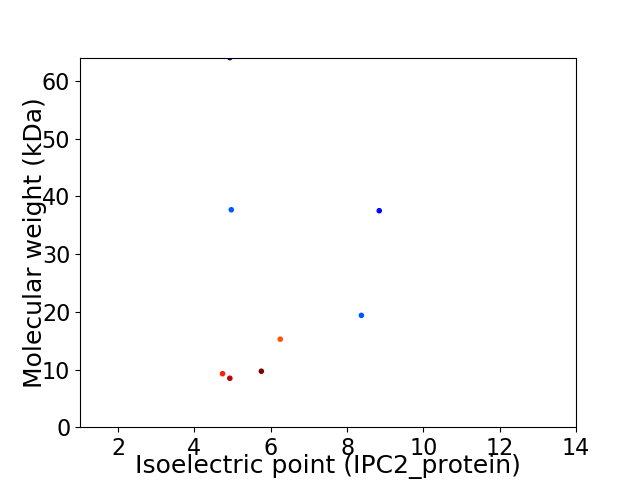
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A4P8W7G9|A0A4P8W7G9_9VIRU Uncharacterized protein OS=Tortoise microvirus 55 OX=2583160 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.7NNVYY6 pKa = 8.78TLYY9 pKa = 9.29NTLSSRR15 pKa = 11.84YY16 pKa = 8.69GEE18 pKa = 4.25VMSFPSDD25 pKa = 2.84AFAVQRR31 pKa = 11.84IRR33 pKa = 11.84EE34 pKa = 4.12AMKK37 pKa = 10.26PEE39 pKa = 3.71MLAEE43 pKa = 4.27IEE45 pKa = 4.2LCNVGFIDD53 pKa = 3.77IDD55 pKa = 3.61TGIVEE60 pKa = 4.03AHH62 pKa = 5.87APVRR66 pKa = 11.84ISLTSPEE73 pKa = 4.41SPVPIDD79 pKa = 3.71NLEE82 pKa = 3.95KK83 pKa = 10.87
MM1 pKa = 7.5KK2 pKa = 10.7NNVYY6 pKa = 8.78TLYY9 pKa = 9.29NTLSSRR15 pKa = 11.84YY16 pKa = 8.69GEE18 pKa = 4.25VMSFPSDD25 pKa = 2.84AFAVQRR31 pKa = 11.84IRR33 pKa = 11.84EE34 pKa = 4.12AMKK37 pKa = 10.26PEE39 pKa = 3.71MLAEE43 pKa = 4.27IEE45 pKa = 4.2LCNVGFIDD53 pKa = 3.77IDD55 pKa = 3.61TGIVEE60 pKa = 4.03AHH62 pKa = 5.87APVRR66 pKa = 11.84ISLTSPEE73 pKa = 4.41SPVPIDD79 pKa = 3.71NLEE82 pKa = 3.95KK83 pKa = 10.87
Molecular weight: 9.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W770|A0A4P8W770_9VIRU Uncharacterized protein OS=Tortoise microvirus 55 OX=2583160 PE=4 SV=1
MM1 pKa = 7.51ISSISLQLLIVTRR14 pKa = 11.84WRR16 pKa = 11.84MSDD19 pKa = 3.26MSCSNPLTINVKK31 pKa = 9.67GRR33 pKa = 11.84AVSVPCRR40 pKa = 11.84YY41 pKa = 10.15CMSCRR46 pKa = 11.84TEE48 pKa = 4.02YY49 pKa = 10.73QSALLFGAKK58 pKa = 9.68HH59 pKa = 5.83EE60 pKa = 5.09LFDD63 pKa = 4.85CYY65 pKa = 10.42RR66 pKa = 11.84AGLGASFVTLTYY78 pKa = 10.87SDD80 pKa = 3.29NCLPEE85 pKa = 4.76NGSLRR90 pKa = 11.84KK91 pKa = 9.53KK92 pKa = 10.45DD93 pKa = 3.61VQNLLKK99 pKa = 10.51NVRR102 pKa = 11.84IQAKK106 pKa = 9.67RR107 pKa = 11.84KK108 pKa = 7.62TNLPPFKK115 pKa = 10.65YY116 pKa = 10.15IMCGEE121 pKa = 4.19YY122 pKa = 10.25GDD124 pKa = 4.21KK125 pKa = 10.43FGRR128 pKa = 11.84PHH130 pKa = 4.73YY131 pKa = 9.98HH132 pKa = 5.82IVFIGLSDD140 pKa = 3.74VLVHH144 pKa = 6.66NFVKK148 pKa = 10.37PLWKK152 pKa = 10.38FGLIDD157 pKa = 3.29IGVLRR162 pKa = 11.84EE163 pKa = 3.78GGLRR167 pKa = 11.84YY168 pKa = 9.55ALKK171 pKa = 10.43YY172 pKa = 7.66CTKK175 pKa = 10.26AARR178 pKa = 11.84GKK180 pKa = 10.15KK181 pKa = 10.14AEE183 pKa = 4.18EE184 pKa = 4.68LYY186 pKa = 11.0DD187 pKa = 3.64NQSIEE192 pKa = 4.29RR193 pKa = 11.84PFICHH198 pKa = 5.72SRR200 pKa = 11.84GIGKK204 pKa = 8.54NWFMKK209 pKa = 10.65NADD212 pKa = 4.98DD213 pKa = 5.48IIAHH217 pKa = 6.03NFTYY221 pKa = 10.71LEE223 pKa = 3.99NGIRR227 pKa = 11.84KK228 pKa = 8.03PVPAYY233 pKa = 9.93FRR235 pKa = 11.84RR236 pKa = 11.84QYY238 pKa = 11.51DD239 pKa = 3.23AFKK242 pKa = 10.04TFDD245 pKa = 3.19VSEE248 pKa = 4.42SIKK251 pKa = 10.67YY252 pKa = 10.17LSEE255 pKa = 3.53EE256 pKa = 4.2ARR258 pKa = 11.84LHH260 pKa = 6.91GYY262 pKa = 10.58DD263 pKa = 5.19NYY265 pKa = 11.19RR266 pKa = 11.84DD267 pKa = 3.24WSKK270 pKa = 11.22QKK272 pKa = 9.69TYY274 pKa = 7.88QTEE277 pKa = 4.25KK278 pKa = 10.76NLTAAARR285 pKa = 11.84ASGIPVDD292 pKa = 4.72DD293 pKa = 3.79SSLRR297 pKa = 11.84LQSGFTANDD306 pKa = 3.06YY307 pKa = 10.93RR308 pKa = 11.84PLVNTLLAPRR318 pKa = 11.84RR319 pKa = 11.84EE320 pKa = 3.94ATTNFKK326 pKa = 10.31EE327 pKa = 4.17
MM1 pKa = 7.51ISSISLQLLIVTRR14 pKa = 11.84WRR16 pKa = 11.84MSDD19 pKa = 3.26MSCSNPLTINVKK31 pKa = 9.67GRR33 pKa = 11.84AVSVPCRR40 pKa = 11.84YY41 pKa = 10.15CMSCRR46 pKa = 11.84TEE48 pKa = 4.02YY49 pKa = 10.73QSALLFGAKK58 pKa = 9.68HH59 pKa = 5.83EE60 pKa = 5.09LFDD63 pKa = 4.85CYY65 pKa = 10.42RR66 pKa = 11.84AGLGASFVTLTYY78 pKa = 10.87SDD80 pKa = 3.29NCLPEE85 pKa = 4.76NGSLRR90 pKa = 11.84KK91 pKa = 9.53KK92 pKa = 10.45DD93 pKa = 3.61VQNLLKK99 pKa = 10.51NVRR102 pKa = 11.84IQAKK106 pKa = 9.67RR107 pKa = 11.84KK108 pKa = 7.62TNLPPFKK115 pKa = 10.65YY116 pKa = 10.15IMCGEE121 pKa = 4.19YY122 pKa = 10.25GDD124 pKa = 4.21KK125 pKa = 10.43FGRR128 pKa = 11.84PHH130 pKa = 4.73YY131 pKa = 9.98HH132 pKa = 5.82IVFIGLSDD140 pKa = 3.74VLVHH144 pKa = 6.66NFVKK148 pKa = 10.37PLWKK152 pKa = 10.38FGLIDD157 pKa = 3.29IGVLRR162 pKa = 11.84EE163 pKa = 3.78GGLRR167 pKa = 11.84YY168 pKa = 9.55ALKK171 pKa = 10.43YY172 pKa = 7.66CTKK175 pKa = 10.26AARR178 pKa = 11.84GKK180 pKa = 10.15KK181 pKa = 10.14AEE183 pKa = 4.18EE184 pKa = 4.68LYY186 pKa = 11.0DD187 pKa = 3.64NQSIEE192 pKa = 4.29RR193 pKa = 11.84PFICHH198 pKa = 5.72SRR200 pKa = 11.84GIGKK204 pKa = 8.54NWFMKK209 pKa = 10.65NADD212 pKa = 4.98DD213 pKa = 5.48IIAHH217 pKa = 6.03NFTYY221 pKa = 10.71LEE223 pKa = 3.99NGIRR227 pKa = 11.84KK228 pKa = 8.03PVPAYY233 pKa = 9.93FRR235 pKa = 11.84RR236 pKa = 11.84QYY238 pKa = 11.51DD239 pKa = 3.23AFKK242 pKa = 10.04TFDD245 pKa = 3.19VSEE248 pKa = 4.42SIKK251 pKa = 10.67YY252 pKa = 10.17LSEE255 pKa = 3.53EE256 pKa = 4.2ARR258 pKa = 11.84LHH260 pKa = 6.91GYY262 pKa = 10.58DD263 pKa = 5.19NYY265 pKa = 11.19RR266 pKa = 11.84DD267 pKa = 3.24WSKK270 pKa = 11.22QKK272 pKa = 9.69TYY274 pKa = 7.88QTEE277 pKa = 4.25KK278 pKa = 10.76NLTAAARR285 pKa = 11.84ASGIPVDD292 pKa = 4.72DD293 pKa = 3.79SSLRR297 pKa = 11.84LQSGFTANDD306 pKa = 3.06YY307 pKa = 10.93RR308 pKa = 11.84PLVNTLLAPRR318 pKa = 11.84RR319 pKa = 11.84EE320 pKa = 3.94ATTNFKK326 pKa = 10.31EE327 pKa = 4.17
Molecular weight: 37.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1791 |
76 |
571 |
223.9 |
25.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.203 ± 1.202 | 1.284 ± 0.518 |
6.253 ± 0.417 | 4.858 ± 0.657 |
4.802 ± 0.641 | 5.36 ± 0.717 |
1.563 ± 0.184 | 5.416 ± 0.791 |
4.69 ± 1.019 | 9.436 ± 1.006 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.127 ± 0.391 | 5.918 ± 0.897 |
4.634 ± 0.608 | 3.573 ± 1.075 |
5.695 ± 0.398 | 9.38 ± 0.741 |
6.477 ± 0.48 | 5.025 ± 0.515 |
0.838 ± 0.22 | 4.467 ± 0.654 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
