
Slackia exigua (strain ATCC 700122 / DSM 15923 / CIP 105133 / JCM 11022 / KCTC 5966 / S-7)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Eggerthellales; Eggerthellaceae; Slackia; Slackia exigua
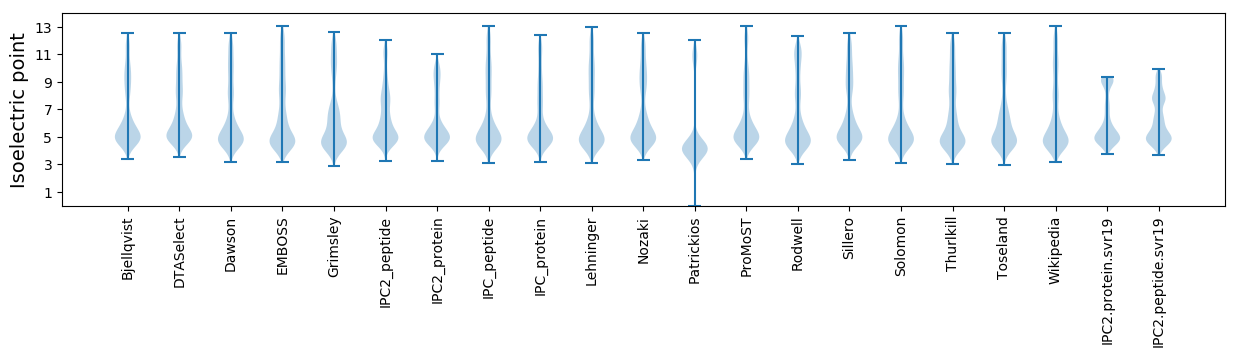
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
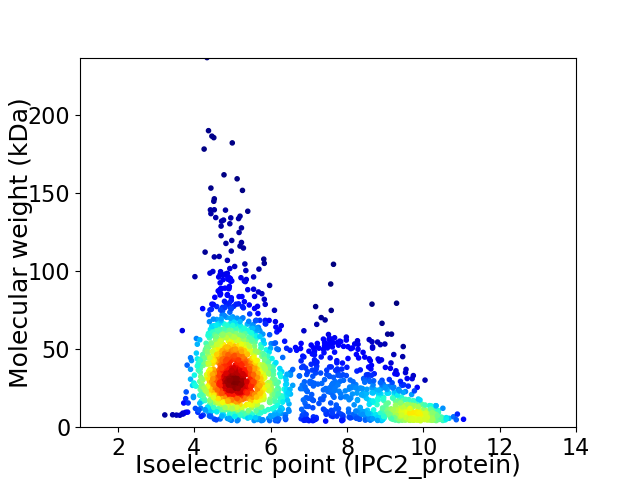
Virtual 2D-PAGE plot for 2027 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D0WHA1|D0WHA1_SLAES Transcriptional regulator TetR family OS=Slackia exigua (strain ATCC 700122 / DSM 15923 / CIP 105133 / JCM 11022 / KCTC 5966 / S-7) OX=649764 GN=HMPREF0762_01133 PE=4 SV=1
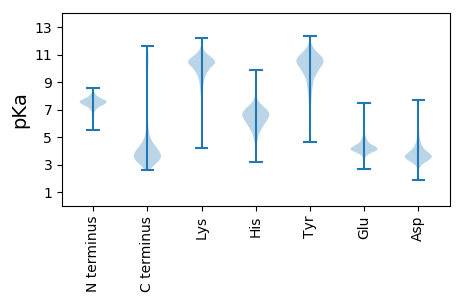
MM1 pKa = 7.6PLKK4 pKa = 8.78EE5 pKa = 4.25TQMTNPTARR14 pKa = 11.84AMSRR18 pKa = 11.84AEE20 pKa = 4.17FLALMAGTAVPAALALSGCSLPAPSDD46 pKa = 3.72LPSGDD51 pKa = 2.92GRR53 pKa = 11.84GIFSPEE59 pKa = 3.8PAPVEE64 pKa = 3.69PLAPVRR70 pKa = 11.84DD71 pKa = 3.82EE72 pKa = 4.1VDD74 pKa = 2.87IPGALEE80 pKa = 4.58DD81 pKa = 4.04VPSSYY86 pKa = 10.82FDD88 pKa = 3.87PSNEE92 pKa = 3.91PGTLEE97 pKa = 4.01RR98 pKa = 11.84FYY100 pKa = 11.29YY101 pKa = 9.18DD102 pKa = 2.89TNTYY106 pKa = 10.88DD107 pKa = 4.3DD108 pKa = 4.18EE109 pKa = 5.91ARR111 pKa = 11.84DD112 pKa = 3.75MEE114 pKa = 4.6KK115 pKa = 10.44FAIVYY120 pKa = 9.44LPYY123 pKa = 10.65GYY125 pKa = 10.77DD126 pKa = 3.42PDD128 pKa = 4.86DD129 pKa = 4.33AEE131 pKa = 3.91TRR133 pKa = 11.84YY134 pKa = 10.33NVLYY138 pKa = 10.57LMHH141 pKa = 6.93GASGSEE147 pKa = 3.64SSYY150 pKa = 10.86FGTPEE155 pKa = 3.74EE156 pKa = 4.42PYY158 pKa = 10.98GDD160 pKa = 4.24KK161 pKa = 10.84FLLDD165 pKa = 3.57SMIAQGDD172 pKa = 4.46MNPMIVVCVSYY183 pKa = 11.15YY184 pKa = 9.81PDD186 pKa = 5.35NIEE189 pKa = 5.07QDD191 pKa = 3.3NDD193 pKa = 3.78DD194 pKa = 4.05YY195 pKa = 12.0DD196 pKa = 3.73AALTKK201 pKa = 10.56RR202 pKa = 11.84FGYY205 pKa = 8.67EE206 pKa = 3.59LRR208 pKa = 11.84DD209 pKa = 3.4LVPQFEE215 pKa = 4.67TRR217 pKa = 11.84YY218 pKa = 8.2NTYY221 pKa = 10.62LEE223 pKa = 4.49SSDD226 pKa = 5.58DD227 pKa = 3.77EE228 pKa = 5.78DD229 pKa = 3.68IVASRR234 pKa = 11.84GHH236 pKa = 6.84RR237 pKa = 11.84IFGGFSMGAVTTWYY251 pKa = 10.86RR252 pKa = 11.84MTDD255 pKa = 2.96SMDD258 pKa = 2.5WFEE261 pKa = 4.97YY262 pKa = 9.05FFPMSGSLFWSSEE275 pKa = 3.81MYY277 pKa = 9.47TEE279 pKa = 4.19RR280 pKa = 11.84RR281 pKa = 11.84SAEE284 pKa = 3.55WTADD288 pKa = 3.6FLVDD292 pKa = 4.97SIDD295 pKa = 3.67DD296 pKa = 3.49QGYY299 pKa = 8.8GLDD302 pKa = 3.87DD303 pKa = 4.04FFVFTTTGSLDD314 pKa = 3.46FALGGLDD321 pKa = 3.53LQVGKK326 pKa = 10.26LRR328 pKa = 11.84KK329 pKa = 9.73HH330 pKa = 6.3PDD332 pKa = 2.99FFVFDD337 pKa = 4.84DD338 pKa = 4.14DD339 pKa = 4.98AEE341 pKa = 4.32NATRR345 pKa = 11.84VAPNAAYY352 pKa = 9.6RR353 pKa = 11.84VAAGYY358 pKa = 10.21SHH360 pKa = 7.68DD361 pKa = 3.69SDD363 pKa = 5.08GRR365 pKa = 11.84DD366 pKa = 3.1TYY368 pKa = 10.86IYY370 pKa = 9.85NCLPIFSEE378 pKa = 4.91LMGG381 pKa = 4.47
MM1 pKa = 7.6PLKK4 pKa = 8.78EE5 pKa = 4.25TQMTNPTARR14 pKa = 11.84AMSRR18 pKa = 11.84AEE20 pKa = 4.17FLALMAGTAVPAALALSGCSLPAPSDD46 pKa = 3.72LPSGDD51 pKa = 2.92GRR53 pKa = 11.84GIFSPEE59 pKa = 3.8PAPVEE64 pKa = 3.69PLAPVRR70 pKa = 11.84DD71 pKa = 3.82EE72 pKa = 4.1VDD74 pKa = 2.87IPGALEE80 pKa = 4.58DD81 pKa = 4.04VPSSYY86 pKa = 10.82FDD88 pKa = 3.87PSNEE92 pKa = 3.91PGTLEE97 pKa = 4.01RR98 pKa = 11.84FYY100 pKa = 11.29YY101 pKa = 9.18DD102 pKa = 2.89TNTYY106 pKa = 10.88DD107 pKa = 4.3DD108 pKa = 4.18EE109 pKa = 5.91ARR111 pKa = 11.84DD112 pKa = 3.75MEE114 pKa = 4.6KK115 pKa = 10.44FAIVYY120 pKa = 9.44LPYY123 pKa = 10.65GYY125 pKa = 10.77DD126 pKa = 3.42PDD128 pKa = 4.86DD129 pKa = 4.33AEE131 pKa = 3.91TRR133 pKa = 11.84YY134 pKa = 10.33NVLYY138 pKa = 10.57LMHH141 pKa = 6.93GASGSEE147 pKa = 3.64SSYY150 pKa = 10.86FGTPEE155 pKa = 3.74EE156 pKa = 4.42PYY158 pKa = 10.98GDD160 pKa = 4.24KK161 pKa = 10.84FLLDD165 pKa = 3.57SMIAQGDD172 pKa = 4.46MNPMIVVCVSYY183 pKa = 11.15YY184 pKa = 9.81PDD186 pKa = 5.35NIEE189 pKa = 5.07QDD191 pKa = 3.3NDD193 pKa = 3.78DD194 pKa = 4.05YY195 pKa = 12.0DD196 pKa = 3.73AALTKK201 pKa = 10.56RR202 pKa = 11.84FGYY205 pKa = 8.67EE206 pKa = 3.59LRR208 pKa = 11.84DD209 pKa = 3.4LVPQFEE215 pKa = 4.67TRR217 pKa = 11.84YY218 pKa = 8.2NTYY221 pKa = 10.62LEE223 pKa = 4.49SSDD226 pKa = 5.58DD227 pKa = 3.77EE228 pKa = 5.78DD229 pKa = 3.68IVASRR234 pKa = 11.84GHH236 pKa = 6.84RR237 pKa = 11.84IFGGFSMGAVTTWYY251 pKa = 10.86RR252 pKa = 11.84MTDD255 pKa = 2.96SMDD258 pKa = 2.5WFEE261 pKa = 4.97YY262 pKa = 9.05FFPMSGSLFWSSEE275 pKa = 3.81MYY277 pKa = 9.47TEE279 pKa = 4.19RR280 pKa = 11.84RR281 pKa = 11.84SAEE284 pKa = 3.55WTADD288 pKa = 3.6FLVDD292 pKa = 4.97SIDD295 pKa = 3.67DD296 pKa = 3.49QGYY299 pKa = 8.8GLDD302 pKa = 3.87DD303 pKa = 4.04FFVFTTTGSLDD314 pKa = 3.46FALGGLDD321 pKa = 3.53LQVGKK326 pKa = 10.26LRR328 pKa = 11.84KK329 pKa = 9.73HH330 pKa = 6.3PDD332 pKa = 2.99FFVFDD337 pKa = 4.84DD338 pKa = 4.14DD339 pKa = 4.98AEE341 pKa = 4.32NATRR345 pKa = 11.84VAPNAAYY352 pKa = 9.6RR353 pKa = 11.84VAAGYY358 pKa = 10.21SHH360 pKa = 7.68DD361 pKa = 3.69SDD363 pKa = 5.08GRR365 pKa = 11.84DD366 pKa = 3.1TYY368 pKa = 10.86IYY370 pKa = 9.85NCLPIFSEE378 pKa = 4.91LMGG381 pKa = 4.47
Molecular weight: 42.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D0WHI0|D0WHI0_SLAES 5-formyltetrahydrofolate cyclo-ligase OS=Slackia exigua (strain ATCC 700122 / DSM 15923 / CIP 105133 / JCM 11022 / KCTC 5966 / S-7) OX=649764 GN=HMPREF0762_01218 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.31QPNKK9 pKa = 7.52RR10 pKa = 11.84HH11 pKa = 5.83RR12 pKa = 11.84AKK14 pKa = 9.65THH16 pKa = 5.05GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.11GGRR28 pKa = 11.84AVLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.06GRR39 pKa = 11.84KK40 pKa = 8.94KK41 pKa = 10.21LTVV44 pKa = 3.1
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.31QPNKK9 pKa = 7.52RR10 pKa = 11.84HH11 pKa = 5.83RR12 pKa = 11.84AKK14 pKa = 9.65THH16 pKa = 5.05GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.11GGRR28 pKa = 11.84AVLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.06GRR39 pKa = 11.84KK40 pKa = 8.94KK41 pKa = 10.21LTVV44 pKa = 3.1
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
632442 |
35 |
2350 |
312.0 |
33.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.087 ± 0.079 | 1.55 ± 0.027 |
6.434 ± 0.052 | 6.147 ± 0.059 |
3.868 ± 0.04 | 8.41 ± 0.05 |
1.936 ± 0.019 | 5.355 ± 0.049 |
3.637 ± 0.045 | 8.722 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.853 ± 0.026 | 2.586 ± 0.029 |
4.246 ± 0.038 | 2.495 ± 0.029 |
6.453 ± 0.077 | 6.413 ± 0.048 |
5.156 ± 0.05 | 7.872 ± 0.051 |
1.037 ± 0.022 | 2.742 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
