
Synechococcus phage ACG-2014f
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Atlauavirus; Synechococcus virus AC2014fSyn7803C8
Average proteome isoelectric point is 5.63
Get precalculated fractions of proteins
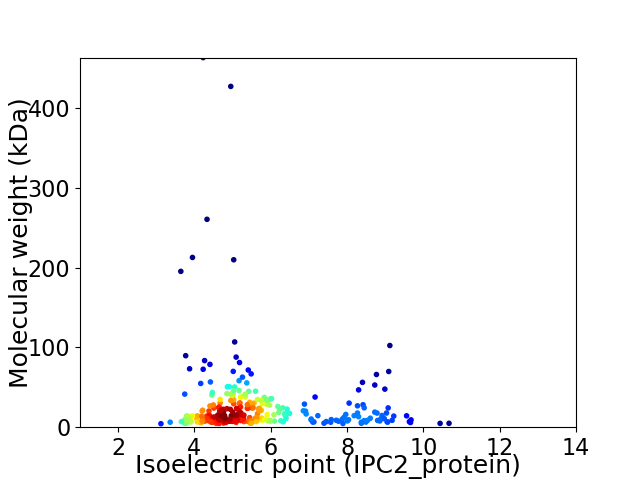
Virtual 2D-PAGE plot for 286 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
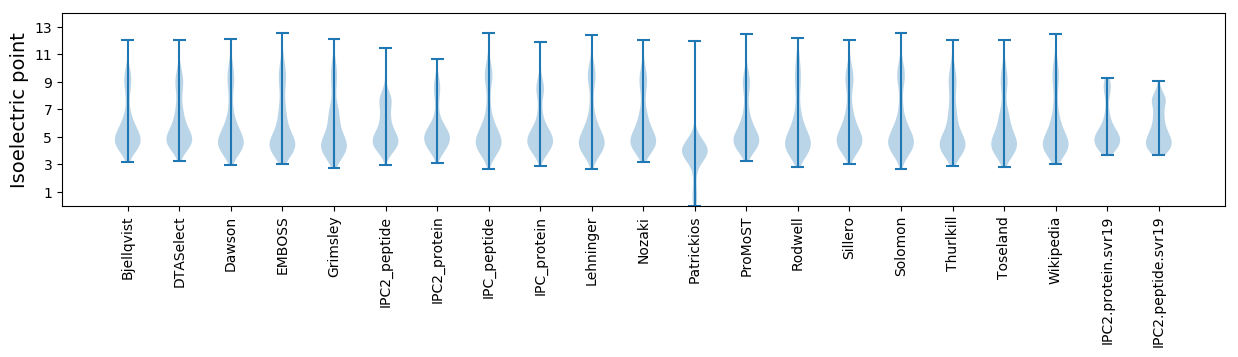
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E3HK52|A0A0E3HK52_9CAUD Uncharacterized protein OS=Synechococcus phage ACG-2014f OX=1493511 GN=Syn7803US3_62 PE=4 SV=1
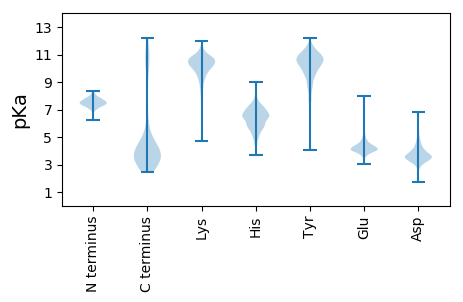
MM1 pKa = 6.87QRR3 pKa = 11.84SGVNYY8 pKa = 9.72KK9 pKa = 10.42VSGLDD14 pKa = 3.63LINRR18 pKa = 11.84DD19 pKa = 3.58TSILNAGDD27 pKa = 3.37VFLVEE32 pKa = 4.8RR33 pKa = 11.84SGTAYY38 pKa = 10.01KK39 pKa = 9.37FTVSLDD45 pKa = 3.13AGGDD49 pKa = 3.38VDD51 pKa = 4.1TSSIQDD57 pKa = 3.54DD58 pKa = 4.0DD59 pKa = 5.77LFVCFIDD66 pKa = 5.03GSEE69 pKa = 3.99KK70 pKa = 10.53KK71 pKa = 10.52INGLEE76 pKa = 4.03FSALVSPNVWEE87 pKa = 4.82TKK89 pKa = 10.52DD90 pKa = 3.31SWTHH94 pKa = 6.37IITDD98 pKa = 4.49IEE100 pKa = 4.47YY101 pKa = 9.19EE102 pKa = 3.99WQSVYY107 pKa = 10.95GYY109 pKa = 10.7DD110 pKa = 3.13HH111 pKa = 7.45DD112 pKa = 4.25GVYY115 pKa = 10.68RR116 pKa = 11.84LDD118 pKa = 3.61GTLIAAAASSTEE130 pKa = 3.96LTTEE134 pKa = 3.94SEE136 pKa = 4.62FVITGNSSTVSAGGIDD152 pKa = 4.43VRR154 pKa = 11.84VAWLSNIGTRR164 pKa = 11.84TDD166 pKa = 3.39LSRR169 pKa = 11.84AFQDD173 pKa = 5.22LPDD176 pKa = 3.68LTLFDD181 pKa = 5.54GYY183 pKa = 10.72WDD185 pKa = 3.68MTNVTNMAQMFYY197 pKa = 11.06NVGPFTADD205 pKa = 2.82VSSWNTSNVTNMSDD219 pKa = 2.71MFYY222 pKa = 10.69RR223 pKa = 11.84SGWGDD228 pKa = 3.12YY229 pKa = 8.89TFNVTGLSSWDD240 pKa = 3.73TTNVTCMSFMFEE252 pKa = 4.89GIDD255 pKa = 3.48LTEE258 pKa = 6.38DD259 pKa = 2.78ITGWDD264 pKa = 3.57VSNVTNMAGMFEE276 pKa = 4.38FGIFNQNIGGWDD288 pKa = 3.51VSNVTNMSYY297 pKa = 9.57MFHH300 pKa = 7.19EE301 pKa = 5.01SNTFNQNIGGWDD313 pKa = 3.5VANVTNMDD321 pKa = 3.07AMFYY325 pKa = 9.8YY326 pKa = 10.86YY327 pKa = 10.65SGGQFAGQFNQNLSGWCVTNIGSKK351 pKa = 10.84PNDD354 pKa = 3.25FDD356 pKa = 4.45TNSGFAGQTALQPQWGTCPP375 pKa = 3.39
MM1 pKa = 6.87QRR3 pKa = 11.84SGVNYY8 pKa = 9.72KK9 pKa = 10.42VSGLDD14 pKa = 3.63LINRR18 pKa = 11.84DD19 pKa = 3.58TSILNAGDD27 pKa = 3.37VFLVEE32 pKa = 4.8RR33 pKa = 11.84SGTAYY38 pKa = 10.01KK39 pKa = 9.37FTVSLDD45 pKa = 3.13AGGDD49 pKa = 3.38VDD51 pKa = 4.1TSSIQDD57 pKa = 3.54DD58 pKa = 4.0DD59 pKa = 5.77LFVCFIDD66 pKa = 5.03GSEE69 pKa = 3.99KK70 pKa = 10.53KK71 pKa = 10.52INGLEE76 pKa = 4.03FSALVSPNVWEE87 pKa = 4.82TKK89 pKa = 10.52DD90 pKa = 3.31SWTHH94 pKa = 6.37IITDD98 pKa = 4.49IEE100 pKa = 4.47YY101 pKa = 9.19EE102 pKa = 3.99WQSVYY107 pKa = 10.95GYY109 pKa = 10.7DD110 pKa = 3.13HH111 pKa = 7.45DD112 pKa = 4.25GVYY115 pKa = 10.68RR116 pKa = 11.84LDD118 pKa = 3.61GTLIAAAASSTEE130 pKa = 3.96LTTEE134 pKa = 3.94SEE136 pKa = 4.62FVITGNSSTVSAGGIDD152 pKa = 4.43VRR154 pKa = 11.84VAWLSNIGTRR164 pKa = 11.84TDD166 pKa = 3.39LSRR169 pKa = 11.84AFQDD173 pKa = 5.22LPDD176 pKa = 3.68LTLFDD181 pKa = 5.54GYY183 pKa = 10.72WDD185 pKa = 3.68MTNVTNMAQMFYY197 pKa = 11.06NVGPFTADD205 pKa = 2.82VSSWNTSNVTNMSDD219 pKa = 2.71MFYY222 pKa = 10.69RR223 pKa = 11.84SGWGDD228 pKa = 3.12YY229 pKa = 8.89TFNVTGLSSWDD240 pKa = 3.73TTNVTCMSFMFEE252 pKa = 4.89GIDD255 pKa = 3.48LTEE258 pKa = 6.38DD259 pKa = 2.78ITGWDD264 pKa = 3.57VSNVTNMAGMFEE276 pKa = 4.38FGIFNQNIGGWDD288 pKa = 3.51VSNVTNMSYY297 pKa = 9.57MFHH300 pKa = 7.19EE301 pKa = 5.01SNTFNQNIGGWDD313 pKa = 3.5VANVTNMDD321 pKa = 3.07AMFYY325 pKa = 9.8YY326 pKa = 10.86YY327 pKa = 10.65SGGQFAGQFNQNLSGWCVTNIGSKK351 pKa = 10.84PNDD354 pKa = 3.25FDD356 pKa = 4.45TNSGFAGQTALQPQWGTCPP375 pKa = 3.39
Molecular weight: 41.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3HGP1|A0A0E3HGP1_9CAUD Uncharacterized protein OS=Synechococcus phage ACG-2014f OX=1493511 GN=Syn7803C12_50 PE=4 SV=1
MM1 pKa = 7.33TKK3 pKa = 9.29ATQRR7 pKa = 11.84PSTQSRR13 pKa = 11.84YY14 pKa = 7.1TSQVAMDD21 pKa = 4.51RR22 pKa = 11.84KK23 pKa = 10.07AAALQRR29 pKa = 11.84AEE31 pKa = 4.2EE32 pKa = 4.0RR33 pKa = 11.84RR34 pKa = 11.84LTGSADD40 pKa = 3.02VDD42 pKa = 3.78QVNRR46 pKa = 11.84LYY48 pKa = 10.68TVTLL52 pKa = 3.77
MM1 pKa = 7.33TKK3 pKa = 9.29ATQRR7 pKa = 11.84PSTQSRR13 pKa = 11.84YY14 pKa = 7.1TSQVAMDD21 pKa = 4.51RR22 pKa = 11.84KK23 pKa = 10.07AAALQRR29 pKa = 11.84AEE31 pKa = 4.2EE32 pKa = 4.0RR33 pKa = 11.84RR34 pKa = 11.84LTGSADD40 pKa = 3.02VDD42 pKa = 3.78QVNRR46 pKa = 11.84LYY48 pKa = 10.68TVTLL52 pKa = 3.77
Molecular weight: 5.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
68578 |
40 |
4225 |
239.8 |
26.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.846 ± 0.212 | 0.949 ± 0.085 |
6.648 ± 0.138 | 6.486 ± 0.28 |
4.445 ± 0.124 | 8.09 ± 0.345 |
1.627 ± 0.127 | 6.483 ± 0.185 |
5.607 ± 0.345 | 7.294 ± 0.139 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.145 ± 0.188 | 5.72 ± 0.169 |
3.963 ± 0.123 | 3.366 ± 0.093 |
4.198 ± 0.141 | 7.301 ± 0.187 |
7.357 ± 0.371 | 7.15 ± 0.234 |
1.111 ± 0.108 | 4.214 ± 0.156 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
