
Oak-Vale virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sripuvirus; Almpiwar sripuvirus
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
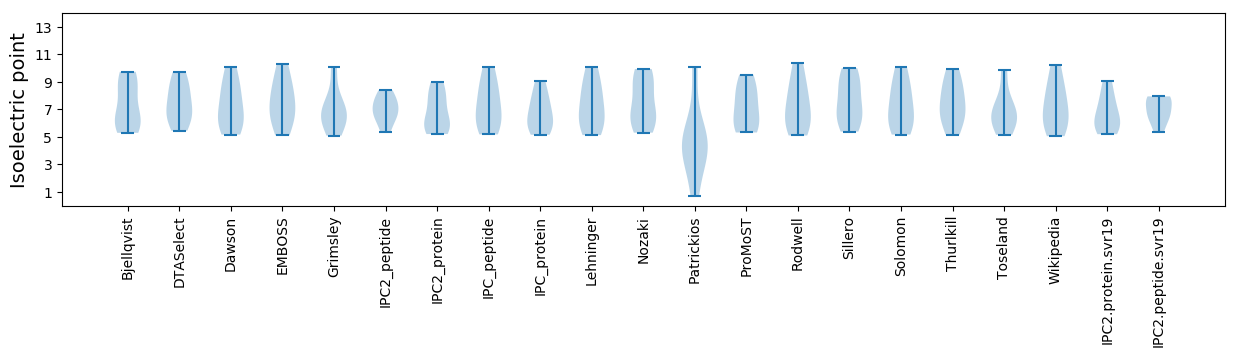
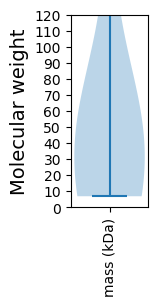

Protein with the lowest isoelectric point:
>tr|G1BWF1|G1BWF1_9RHAB Putative SH protein OS=Oak-Vale virus OX=318852 GN=SH PE=4 SV=1
MM1 pKa = 7.27NLSPEE6 pKa = 4.11QLTEE10 pKa = 3.82LSSFVTQHH18 pKa = 5.6QLEE21 pKa = 4.55DD22 pKa = 4.66DD23 pKa = 3.96EE24 pKa = 7.18DD25 pKa = 4.17SLAEE29 pKa = 4.48KK30 pKa = 10.0VAKK33 pKa = 9.98SCSISEE39 pKa = 4.33KK40 pKa = 10.36VFPPNPSDD48 pKa = 3.64TEE50 pKa = 4.29GEE52 pKa = 4.01IMDD55 pKa = 3.96YY56 pKa = 10.59QSSGEE61 pKa = 4.11SDD63 pKa = 3.41GQANPSLEE71 pKa = 3.92WDD73 pKa = 3.53HH74 pKa = 6.99SVLARR79 pKa = 11.84AGDD82 pKa = 3.9SDD84 pKa = 3.55HH85 pKa = 6.48TRR87 pKa = 11.84PGGSLPTPGPSGTGGPSGLIGGGSGLIAIPQTPPAIIHH125 pKa = 5.66QPPVDD130 pKa = 4.11ASPPLIPAIPSQVTLTLADD149 pKa = 3.5AMSLFNSVLAKK160 pKa = 10.2SDD162 pKa = 3.3EE163 pKa = 4.14GSFYY167 pKa = 11.27LEE169 pKa = 4.35AGQIKK174 pKa = 10.26YY175 pKa = 10.38LSKK178 pKa = 10.78KK179 pKa = 9.01KK180 pKa = 9.62QQNLTRR186 pKa = 11.84SVLSGPDD193 pKa = 3.16SPLPGMEE200 pKa = 4.76SSPKK204 pKa = 8.4PQAVSRR210 pKa = 11.84GCQTTSSCIATSKK223 pKa = 10.63PPTPPPRR230 pKa = 11.84RR231 pKa = 11.84TLAANFSTPLAPPEE245 pKa = 4.0KK246 pKa = 9.67SKK248 pKa = 11.48SSGEE252 pKa = 4.07RR253 pKa = 11.84EE254 pKa = 3.99AVIPPTGIAWNPFCIFPEE272 pKa = 4.28PPGNKK277 pKa = 9.17DD278 pKa = 3.07GKK280 pKa = 9.26EE281 pKa = 3.8ASPEE285 pKa = 3.92STHH288 pKa = 8.14IEE290 pKa = 3.9EE291 pKa = 5.14HH292 pKa = 6.62PPGDD296 pKa = 3.23TGGNPFVQNPEE307 pKa = 3.51GAQIVDD313 pKa = 4.22RR314 pKa = 11.84PTAPPLGRR322 pKa = 11.84DD323 pKa = 3.64GAISNYY329 pKa = 10.07AGSPPNVPRR338 pKa = 11.84YY339 pKa = 10.19SLTEE343 pKa = 3.75LRR345 pKa = 11.84VKK347 pKa = 10.31SEE349 pKa = 3.84KK350 pKa = 10.48GFKK353 pKa = 10.04IKK355 pKa = 10.5GLNKK359 pKa = 9.83ILTIYY364 pKa = 10.32DD365 pKa = 3.72FRR367 pKa = 11.84WDD369 pKa = 3.72EE370 pKa = 4.21MTSHH374 pKa = 6.22QGKK377 pKa = 9.24FPLKK381 pKa = 9.36FWLRR385 pKa = 11.84FANII389 pKa = 3.42
MM1 pKa = 7.27NLSPEE6 pKa = 4.11QLTEE10 pKa = 3.82LSSFVTQHH18 pKa = 5.6QLEE21 pKa = 4.55DD22 pKa = 4.66DD23 pKa = 3.96EE24 pKa = 7.18DD25 pKa = 4.17SLAEE29 pKa = 4.48KK30 pKa = 10.0VAKK33 pKa = 9.98SCSISEE39 pKa = 4.33KK40 pKa = 10.36VFPPNPSDD48 pKa = 3.64TEE50 pKa = 4.29GEE52 pKa = 4.01IMDD55 pKa = 3.96YY56 pKa = 10.59QSSGEE61 pKa = 4.11SDD63 pKa = 3.41GQANPSLEE71 pKa = 3.92WDD73 pKa = 3.53HH74 pKa = 6.99SVLARR79 pKa = 11.84AGDD82 pKa = 3.9SDD84 pKa = 3.55HH85 pKa = 6.48TRR87 pKa = 11.84PGGSLPTPGPSGTGGPSGLIGGGSGLIAIPQTPPAIIHH125 pKa = 5.66QPPVDD130 pKa = 4.11ASPPLIPAIPSQVTLTLADD149 pKa = 3.5AMSLFNSVLAKK160 pKa = 10.2SDD162 pKa = 3.3EE163 pKa = 4.14GSFYY167 pKa = 11.27LEE169 pKa = 4.35AGQIKK174 pKa = 10.26YY175 pKa = 10.38LSKK178 pKa = 10.78KK179 pKa = 9.01KK180 pKa = 9.62QQNLTRR186 pKa = 11.84SVLSGPDD193 pKa = 3.16SPLPGMEE200 pKa = 4.76SSPKK204 pKa = 8.4PQAVSRR210 pKa = 11.84GCQTTSSCIATSKK223 pKa = 10.63PPTPPPRR230 pKa = 11.84RR231 pKa = 11.84TLAANFSTPLAPPEE245 pKa = 4.0KK246 pKa = 9.67SKK248 pKa = 11.48SSGEE252 pKa = 4.07RR253 pKa = 11.84EE254 pKa = 3.99AVIPPTGIAWNPFCIFPEE272 pKa = 4.28PPGNKK277 pKa = 9.17DD278 pKa = 3.07GKK280 pKa = 9.26EE281 pKa = 3.8ASPEE285 pKa = 3.92STHH288 pKa = 8.14IEE290 pKa = 3.9EE291 pKa = 5.14HH292 pKa = 6.62PPGDD296 pKa = 3.23TGGNPFVQNPEE307 pKa = 3.51GAQIVDD313 pKa = 4.22RR314 pKa = 11.84PTAPPLGRR322 pKa = 11.84DD323 pKa = 3.64GAISNYY329 pKa = 10.07AGSPPNVPRR338 pKa = 11.84YY339 pKa = 10.19SLTEE343 pKa = 3.75LRR345 pKa = 11.84VKK347 pKa = 10.31SEE349 pKa = 3.84KK350 pKa = 10.48GFKK353 pKa = 10.04IKK355 pKa = 10.5GLNKK359 pKa = 9.83ILTIYY364 pKa = 10.32DD365 pKa = 3.72FRR367 pKa = 11.84WDD369 pKa = 3.72EE370 pKa = 4.21MTSHH374 pKa = 6.22QGKK377 pKa = 9.24FPLKK381 pKa = 9.36FWLRR385 pKa = 11.84FANII389 pKa = 3.42
Molecular weight: 41.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G1BWE8|G1BWE8_9RHAB Putative phosphoprotein OS=Oak-Vale virus OX=318852 GN=P PE=4 SV=1
MM1 pKa = 7.43NLLKK5 pKa = 10.47FFSSGDD11 pKa = 3.61PLAVSSAARR20 pKa = 11.84RR21 pKa = 11.84NARR24 pKa = 11.84ISYY27 pKa = 10.35SLDD30 pKa = 2.75ITLDD34 pKa = 3.62LSRR37 pKa = 11.84INVSLDD43 pKa = 3.06HH44 pKa = 6.83KK45 pKa = 11.19NVIEE49 pKa = 4.54AIKK52 pKa = 10.42RR53 pKa = 11.84VISPPLEE60 pKa = 3.86IRR62 pKa = 11.84SLICYY67 pKa = 9.36LLTSIRR73 pKa = 11.84TFRR76 pKa = 11.84VIKK79 pKa = 10.79GEE81 pKa = 3.9TRR83 pKa = 11.84TVGIKK88 pKa = 9.48WEE90 pKa = 4.16LSKK93 pKa = 10.7LTVPTDD99 pKa = 3.64FPEE102 pKa = 4.98RR103 pKa = 11.84FDD105 pKa = 4.05KK106 pKa = 10.74RR107 pKa = 11.84WIVHH111 pKa = 6.25RR112 pKa = 11.84EE113 pKa = 4.0GILPGFDD120 pKa = 3.0RR121 pKa = 11.84PIMYY125 pKa = 9.62KK126 pKa = 9.14VHH128 pKa = 6.78ICIEE132 pKa = 4.35PGPGGTAPDD141 pKa = 4.16FKK143 pKa = 10.62ILSLMSEE150 pKa = 4.14EE151 pKa = 3.96EE152 pKa = 3.83RR153 pKa = 11.84RR154 pKa = 11.84SNFTQRR160 pKa = 11.84DD161 pKa = 3.78RR162 pKa = 11.84GWVFNN167 pKa = 4.48
MM1 pKa = 7.43NLLKK5 pKa = 10.47FFSSGDD11 pKa = 3.61PLAVSSAARR20 pKa = 11.84RR21 pKa = 11.84NARR24 pKa = 11.84ISYY27 pKa = 10.35SLDD30 pKa = 2.75ITLDD34 pKa = 3.62LSRR37 pKa = 11.84INVSLDD43 pKa = 3.06HH44 pKa = 6.83KK45 pKa = 11.19NVIEE49 pKa = 4.54AIKK52 pKa = 10.42RR53 pKa = 11.84VISPPLEE60 pKa = 3.86IRR62 pKa = 11.84SLICYY67 pKa = 9.36LLTSIRR73 pKa = 11.84TFRR76 pKa = 11.84VIKK79 pKa = 10.79GEE81 pKa = 3.9TRR83 pKa = 11.84TVGIKK88 pKa = 9.48WEE90 pKa = 4.16LSKK93 pKa = 10.7LTVPTDD99 pKa = 3.64FPEE102 pKa = 4.98RR103 pKa = 11.84FDD105 pKa = 4.05KK106 pKa = 10.74RR107 pKa = 11.84WIVHH111 pKa = 6.25RR112 pKa = 11.84EE113 pKa = 4.0GILPGFDD120 pKa = 3.0RR121 pKa = 11.84PIMYY125 pKa = 9.62KK126 pKa = 9.14VHH128 pKa = 6.78ICIEE132 pKa = 4.35PGPGGTAPDD141 pKa = 4.16FKK143 pKa = 10.62ILSLMSEE150 pKa = 4.14EE151 pKa = 3.96EE152 pKa = 3.83RR153 pKa = 11.84RR154 pKa = 11.84SNFTQRR160 pKa = 11.84DD161 pKa = 3.78RR162 pKa = 11.84GWVFNN167 pKa = 4.48
Molecular weight: 19.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3738 |
60 |
2051 |
534.0 |
60.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.735 ± 0.874 | 2.06 ± 0.259 |
5.056 ± 0.244 | 6.26 ± 0.203 |
4.334 ± 0.284 | 7.143 ± 0.445 |
2.327 ± 0.304 | 7.598 ± 0.523 |
6.18 ± 1.047 | 9.524 ± 0.823 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.622 ± 0.378 | 4.361 ± 0.562 |
5.297 ± 1.453 | 3.264 ± 0.544 |
5.752 ± 0.911 | 7.571 ± 0.945 |
5.805 ± 0.332 | 5.778 ± 0.455 |
1.418 ± 0.132 | 2.916 ± 0.427 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
