
Wuhan Insect virus 6
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus; Wuhan 6 insect cytorhabdovirus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
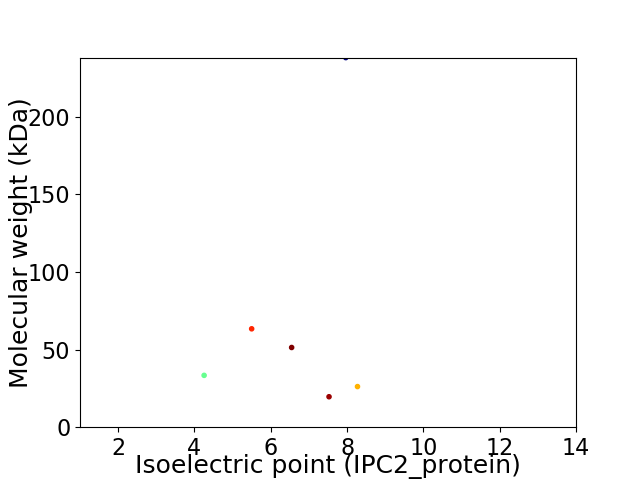
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KXN7|A0A0B5KXN7_9RHAB 4b protein OS=Wuhan Insect virus 6 OX=1608111 GN=4b PE=4 SV=1
MM1 pKa = 7.65ASSSDD6 pKa = 3.5VPNYY10 pKa = 10.77SDD12 pKa = 4.2VGEE15 pKa = 4.49PLDD18 pKa = 3.83NLNYY22 pKa = 10.41TLFNDD27 pKa = 5.29DD28 pKa = 5.83DD29 pKa = 4.68DD30 pKa = 6.87LEE32 pKa = 6.4DD33 pKa = 5.35DD34 pKa = 4.34SPIVCTTTIPVSSSEE49 pKa = 4.4VVVSTHH55 pKa = 6.76KK56 pKa = 10.58EE57 pKa = 4.0SPNLKK62 pKa = 10.19DD63 pKa = 5.89ALDD66 pKa = 4.21EE67 pKa = 4.18LQNMCMEE74 pKa = 4.52KK75 pKa = 10.78GIVCTLAMEE84 pKa = 4.57NQVKK88 pKa = 9.64MISSSEE94 pKa = 4.11EE95 pKa = 3.65IYY97 pKa = 10.55KK98 pKa = 10.35SHH100 pKa = 6.1LTWYY104 pKa = 10.37LRR106 pKa = 11.84GIAMANNTQLLPSIHH121 pKa = 7.34DD122 pKa = 3.97SIGVLKK128 pKa = 10.98SEE130 pKa = 4.65IKK132 pKa = 9.26FTQLSTNNLNNASAKK147 pKa = 9.39LTGAVQEE154 pKa = 4.03ITRR157 pKa = 11.84EE158 pKa = 3.95IRR160 pKa = 11.84TSTEE164 pKa = 3.75MINDD168 pKa = 3.35NFKK171 pKa = 10.05TVLLQISEE179 pKa = 4.32NQVLPSPNSDD189 pKa = 3.49KK190 pKa = 11.49EE191 pKa = 3.64EE192 pKa = 4.0STYY195 pKa = 11.5VKK197 pKa = 10.32VDD199 pKa = 4.42PIYY202 pKa = 10.88QSDD205 pKa = 4.12TVVTSQEE212 pKa = 4.14FTQSEE217 pKa = 4.6SSPDD221 pKa = 3.35AKK223 pKa = 10.69FSVADD228 pKa = 3.52KK229 pKa = 11.42SDD231 pKa = 3.6FNKK234 pKa = 9.93MKK236 pKa = 10.52SAFLISLGMDD246 pKa = 3.77PDD248 pKa = 4.36LLRR251 pKa = 11.84DD252 pKa = 4.16LPDD255 pKa = 4.14GDD257 pKa = 5.57LDD259 pKa = 5.85LFLSDD264 pKa = 4.41NDD266 pKa = 3.63FKK268 pKa = 11.21EE269 pKa = 4.18FRR271 pKa = 11.84SMKK274 pKa = 9.0LTNALKK280 pKa = 10.58RR281 pKa = 11.84EE282 pKa = 4.0LLQTLMKK289 pKa = 10.52EE290 pKa = 4.59FEE292 pKa = 4.39TKK294 pKa = 10.6LDD296 pKa = 4.12DD297 pKa = 4.51GLL299 pKa = 5.17
MM1 pKa = 7.65ASSSDD6 pKa = 3.5VPNYY10 pKa = 10.77SDD12 pKa = 4.2VGEE15 pKa = 4.49PLDD18 pKa = 3.83NLNYY22 pKa = 10.41TLFNDD27 pKa = 5.29DD28 pKa = 5.83DD29 pKa = 4.68DD30 pKa = 6.87LEE32 pKa = 6.4DD33 pKa = 5.35DD34 pKa = 4.34SPIVCTTTIPVSSSEE49 pKa = 4.4VVVSTHH55 pKa = 6.76KK56 pKa = 10.58EE57 pKa = 4.0SPNLKK62 pKa = 10.19DD63 pKa = 5.89ALDD66 pKa = 4.21EE67 pKa = 4.18LQNMCMEE74 pKa = 4.52KK75 pKa = 10.78GIVCTLAMEE84 pKa = 4.57NQVKK88 pKa = 9.64MISSSEE94 pKa = 4.11EE95 pKa = 3.65IYY97 pKa = 10.55KK98 pKa = 10.35SHH100 pKa = 6.1LTWYY104 pKa = 10.37LRR106 pKa = 11.84GIAMANNTQLLPSIHH121 pKa = 7.34DD122 pKa = 3.97SIGVLKK128 pKa = 10.98SEE130 pKa = 4.65IKK132 pKa = 9.26FTQLSTNNLNNASAKK147 pKa = 9.39LTGAVQEE154 pKa = 4.03ITRR157 pKa = 11.84EE158 pKa = 3.95IRR160 pKa = 11.84TSTEE164 pKa = 3.75MINDD168 pKa = 3.35NFKK171 pKa = 10.05TVLLQISEE179 pKa = 4.32NQVLPSPNSDD189 pKa = 3.49KK190 pKa = 11.49EE191 pKa = 3.64EE192 pKa = 4.0STYY195 pKa = 11.5VKK197 pKa = 10.32VDD199 pKa = 4.42PIYY202 pKa = 10.88QSDD205 pKa = 4.12TVVTSQEE212 pKa = 4.14FTQSEE217 pKa = 4.6SSPDD221 pKa = 3.35AKK223 pKa = 10.69FSVADD228 pKa = 3.52KK229 pKa = 11.42SDD231 pKa = 3.6FNKK234 pKa = 9.93MKK236 pKa = 10.52SAFLISLGMDD246 pKa = 3.77PDD248 pKa = 4.36LLRR251 pKa = 11.84DD252 pKa = 4.16LPDD255 pKa = 4.14GDD257 pKa = 5.57LDD259 pKa = 5.85LFLSDD264 pKa = 4.41NDD266 pKa = 3.63FKK268 pKa = 11.21EE269 pKa = 4.18FRR271 pKa = 11.84SMKK274 pKa = 9.0LTNALKK280 pKa = 10.58RR281 pKa = 11.84EE282 pKa = 4.0LLQTLMKK289 pKa = 10.52EE290 pKa = 4.59FEE292 pKa = 4.39TKK294 pKa = 10.6LDD296 pKa = 4.12DD297 pKa = 4.51GLL299 pKa = 5.17
Molecular weight: 33.41 kDa
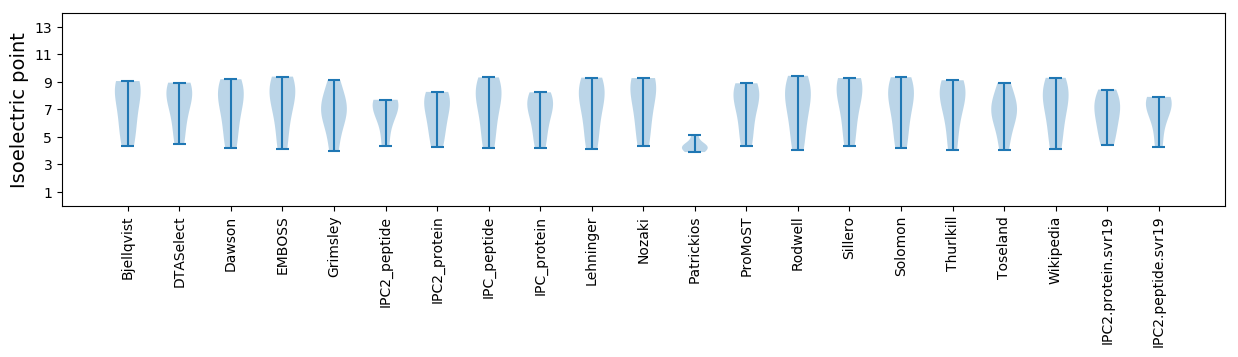
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KXN7|A0A0B5KXN7_9RHAB 4b protein OS=Wuhan Insect virus 6 OX=1608111 GN=4b PE=4 SV=1
MM1 pKa = 7.53SLTFRR6 pKa = 11.84DD7 pKa = 3.81TVKK10 pKa = 11.06YY11 pKa = 9.84EE12 pKa = 4.03LVVSEE17 pKa = 4.17NGGVVSLTKK26 pKa = 10.25RR27 pKa = 11.84LNFFRR32 pKa = 11.84EE33 pKa = 4.21WMTKK37 pKa = 8.71GHH39 pKa = 6.69DD40 pKa = 3.31NVRR43 pKa = 11.84IKK45 pKa = 11.14NLIVKK50 pKa = 8.7YY51 pKa = 10.18NSRR54 pKa = 11.84AGPLGSGTIITRR66 pKa = 11.84VRR68 pKa = 11.84DD69 pKa = 3.51NRR71 pKa = 11.84IDD73 pKa = 3.62SDD75 pKa = 4.03LSEE78 pKa = 5.17SSDD81 pKa = 3.41NTLCTAEE88 pKa = 4.44FPVSEE93 pKa = 4.33SVILRR98 pKa = 11.84WEE100 pKa = 4.53SNVWLYY106 pKa = 11.13KK107 pKa = 10.63GDD109 pKa = 4.15LNNVDD114 pKa = 3.97NPPLIFEE121 pKa = 4.87IDD123 pKa = 3.51LVACNMTLGNSVGQFAITCEE143 pKa = 3.98MAFSKK148 pKa = 11.18VMDD151 pKa = 4.65KK152 pKa = 10.52IRR154 pKa = 11.84YY155 pKa = 7.42KK156 pKa = 10.91NPSAKK161 pKa = 10.55LLDD164 pKa = 3.73TSSVKK169 pKa = 10.49GKK171 pKa = 10.2NIEE174 pKa = 4.73HH175 pKa = 5.98ITKK178 pKa = 10.32FSLTDD183 pKa = 3.09KK184 pKa = 10.65GLEE187 pKa = 3.88NGAQLRR193 pKa = 11.84ARR195 pKa = 11.84QTTHH199 pKa = 6.79RR200 pKa = 11.84ADD202 pKa = 5.16GIRR205 pKa = 11.84KK206 pKa = 7.99TIDD209 pKa = 3.22PGLSSNKK216 pKa = 9.4EE217 pKa = 4.01VLDD220 pKa = 4.35SIATKK225 pKa = 10.32RR226 pKa = 11.84ALGYY230 pKa = 10.81HH231 pKa = 6.8PDD233 pKa = 3.51KK234 pKa = 11.41
MM1 pKa = 7.53SLTFRR6 pKa = 11.84DD7 pKa = 3.81TVKK10 pKa = 11.06YY11 pKa = 9.84EE12 pKa = 4.03LVVSEE17 pKa = 4.17NGGVVSLTKK26 pKa = 10.25RR27 pKa = 11.84LNFFRR32 pKa = 11.84EE33 pKa = 4.21WMTKK37 pKa = 8.71GHH39 pKa = 6.69DD40 pKa = 3.31NVRR43 pKa = 11.84IKK45 pKa = 11.14NLIVKK50 pKa = 8.7YY51 pKa = 10.18NSRR54 pKa = 11.84AGPLGSGTIITRR66 pKa = 11.84VRR68 pKa = 11.84DD69 pKa = 3.51NRR71 pKa = 11.84IDD73 pKa = 3.62SDD75 pKa = 4.03LSEE78 pKa = 5.17SSDD81 pKa = 3.41NTLCTAEE88 pKa = 4.44FPVSEE93 pKa = 4.33SVILRR98 pKa = 11.84WEE100 pKa = 4.53SNVWLYY106 pKa = 11.13KK107 pKa = 10.63GDD109 pKa = 4.15LNNVDD114 pKa = 3.97NPPLIFEE121 pKa = 4.87IDD123 pKa = 3.51LVACNMTLGNSVGQFAITCEE143 pKa = 3.98MAFSKK148 pKa = 11.18VMDD151 pKa = 4.65KK152 pKa = 10.52IRR154 pKa = 11.84YY155 pKa = 7.42KK156 pKa = 10.91NPSAKK161 pKa = 10.55LLDD164 pKa = 3.73TSSVKK169 pKa = 10.49GKK171 pKa = 10.2NIEE174 pKa = 4.73HH175 pKa = 5.98ITKK178 pKa = 10.32FSLTDD183 pKa = 3.09KK184 pKa = 10.65GLEE187 pKa = 3.88NGAQLRR193 pKa = 11.84ARR195 pKa = 11.84QTTHH199 pKa = 6.79RR200 pKa = 11.84ADD202 pKa = 5.16GIRR205 pKa = 11.84KK206 pKa = 7.99TIDD209 pKa = 3.22PGLSSNKK216 pKa = 9.4EE217 pKa = 4.01VLDD220 pKa = 4.35SIATKK225 pKa = 10.32RR226 pKa = 11.84ALGYY230 pKa = 10.81HH231 pKa = 6.8PDD233 pKa = 3.51KK234 pKa = 11.41
Molecular weight: 26.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3805 |
173 |
2079 |
634.2 |
72.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.047 ± 0.692 | 1.866 ± 0.325 |
6.702 ± 0.382 | 5.256 ± 0.729 |
3.233 ± 0.241 | 5.309 ± 0.37 |
2.102 ± 0.225 | 6.649 ± 0.26 |
6.57 ± 0.614 | 10.066 ± 0.232 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.679 ± 0.346 | 5.23 ± 0.519 |
4.258 ± 0.243 | 2.444 ± 0.269 |
5.361 ± 0.778 | 8.988 ± 0.351 |
6.439 ± 0.188 | 6.439 ± 0.137 |
1.603 ± 0.276 | 3.758 ± 0.386 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
