
Coprococcus sp. CAG:782
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Coprococcus; environmental samples
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
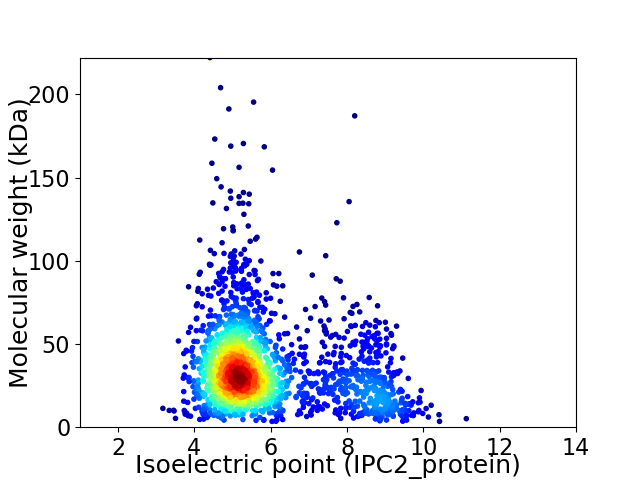
Virtual 2D-PAGE plot for 1959 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5JE35|R5JE35_9FIRM Uncharacterized protein OS=Coprococcus sp. CAG:782 OX=1262863 GN=BN781_01262 PE=4 SV=1
MM1 pKa = 7.43KK2 pKa = 10.33KK3 pKa = 10.21SFVRR7 pKa = 11.84RR8 pKa = 11.84AVALSMIGVMMLSGAATGCGKK29 pKa = 10.6KK30 pKa = 10.15KK31 pKa = 10.26VDD33 pKa = 3.94YY34 pKa = 11.41NIDD37 pKa = 3.78DD38 pKa = 4.02EE39 pKa = 5.2NGGSEE44 pKa = 4.11NGGSGDD50 pKa = 4.11SGEE53 pKa = 4.62LRR55 pKa = 11.84AKK57 pKa = 10.26VKK59 pKa = 10.73APEE62 pKa = 4.67SYY64 pKa = 10.17TGDD67 pKa = 3.29IPVGNSGLSSIKK79 pKa = 10.33INAEE83 pKa = 3.71KK84 pKa = 10.52IEE86 pKa = 4.21IPNASTMSVVYY97 pKa = 9.47CEE99 pKa = 4.12PLSYY103 pKa = 11.2DD104 pKa = 3.16SDD106 pKa = 4.13YY107 pKa = 11.61KK108 pKa = 10.35KK109 pKa = 9.07TLAEE113 pKa = 4.16SFFDD117 pKa = 3.62KK118 pKa = 11.11SKK120 pKa = 11.28GIYY123 pKa = 9.67KK124 pKa = 9.99YY125 pKa = 10.58DD126 pKa = 3.39YY127 pKa = 8.38EE128 pKa = 4.69KK129 pKa = 10.62PIRR132 pKa = 11.84EE133 pKa = 4.19DD134 pKa = 3.22VQKK137 pKa = 10.99EE138 pKa = 3.89LDD140 pKa = 3.76VYY142 pKa = 11.2QAMLDD147 pKa = 3.81DD148 pKa = 5.28ANAAGDD154 pKa = 3.91TEE156 pKa = 4.14NAQWIGDD163 pKa = 3.94TVNSLKK169 pKa = 10.65SDD171 pKa = 3.67LAKK174 pKa = 10.31ATDD177 pKa = 3.35EE178 pKa = 4.35RR179 pKa = 11.84EE180 pKa = 4.4PVGDD184 pKa = 4.04TYY186 pKa = 11.71DD187 pKa = 3.35SDD189 pKa = 4.86SYY191 pKa = 10.98IGYY194 pKa = 9.8IGDD197 pKa = 3.55QLYY200 pKa = 10.49EE201 pKa = 4.24LDD203 pKa = 4.08MYY205 pKa = 11.36GDD207 pKa = 3.83SSGVNGSASIAPYY220 pKa = 10.01PYY222 pKa = 9.05ITIKK226 pKa = 10.56DD227 pKa = 3.65IRR229 pKa = 11.84PHH231 pKa = 6.56DD232 pKa = 3.86DD233 pKa = 3.81CYY235 pKa = 11.28NVSYY239 pKa = 11.3YY240 pKa = 11.06GDD242 pKa = 4.56DD243 pKa = 3.66YY244 pKa = 11.95NSYY247 pKa = 11.13DD248 pKa = 5.23LEE250 pKa = 4.43NQSDD254 pKa = 3.86MTKK257 pKa = 10.92DD258 pKa = 3.32EE259 pKa = 4.35AVSYY263 pKa = 8.11ATSYY267 pKa = 10.94LADD270 pKa = 4.46LGFKK274 pKa = 10.61DD275 pKa = 5.93AMLKK279 pKa = 10.24SCSPLIWDD287 pKa = 4.0YY288 pKa = 11.82YY289 pKa = 11.14DD290 pKa = 3.47YY291 pKa = 11.68AGNTLAEE298 pKa = 4.47EE299 pKa = 4.39CDD301 pKa = 3.24GWSITFVKK309 pKa = 10.63SVDD312 pKa = 3.41NVPIYY317 pKa = 11.08SPDD320 pKa = 3.18VWNLDD325 pKa = 3.62YY326 pKa = 11.52LDD328 pKa = 4.7TNNVWYY334 pKa = 6.89TTSDD338 pKa = 3.1QTINLSIYY346 pKa = 10.02NGSAFQVNINYY357 pKa = 10.14SLNTIKK363 pKa = 10.58EE364 pKa = 4.15DD365 pKa = 3.61SDD367 pKa = 4.35VEE369 pKa = 4.16LLSWDD374 pKa = 4.68EE375 pKa = 4.06ILKK378 pKa = 10.35SAQEE382 pKa = 4.52KK383 pKa = 10.38IPEE386 pKa = 4.24FYY388 pKa = 10.66KK389 pKa = 10.9DD390 pKa = 3.36NKK392 pKa = 9.7TSYY395 pKa = 11.33LDD397 pKa = 3.93LEE399 pKa = 4.21FDD401 pKa = 3.96KK402 pKa = 11.65VKK404 pKa = 9.77LTYY407 pKa = 10.29FRR409 pKa = 11.84VKK411 pKa = 10.97DD412 pKa = 3.61EE413 pKa = 4.56DD414 pKa = 3.66TTGQYY419 pKa = 11.65KK420 pKa = 10.53LIPVWAFISTSYY432 pKa = 11.19GDD434 pKa = 3.95EE435 pKa = 4.29EE436 pKa = 6.54GDD438 pKa = 3.38TSTEE442 pKa = 3.74EE443 pKa = 3.97DD444 pKa = 3.76TEE446 pKa = 4.19EE447 pKa = 4.16DD448 pKa = 3.89TEE450 pKa = 4.63DD451 pKa = 3.52SAFEE455 pKa = 4.23DD456 pKa = 4.04SYY458 pKa = 10.4PTQLILINAMDD469 pKa = 4.47GSLIDD474 pKa = 3.61VASQLEE480 pKa = 4.48GEE482 pKa = 4.93SNDD485 pKa = 3.59SDD487 pKa = 4.58DD488 pKa = 4.08VTSSTISDD496 pKa = 3.71DD497 pKa = 3.48GSFAVEE503 pKa = 4.18KK504 pKa = 10.52EE505 pKa = 3.77VDD507 pKa = 3.36EE508 pKa = 5.91GDD510 pKa = 3.52IQQ512 pKa = 4.11
MM1 pKa = 7.43KK2 pKa = 10.33KK3 pKa = 10.21SFVRR7 pKa = 11.84RR8 pKa = 11.84AVALSMIGVMMLSGAATGCGKK29 pKa = 10.6KK30 pKa = 10.15KK31 pKa = 10.26VDD33 pKa = 3.94YY34 pKa = 11.41NIDD37 pKa = 3.78DD38 pKa = 4.02EE39 pKa = 5.2NGGSEE44 pKa = 4.11NGGSGDD50 pKa = 4.11SGEE53 pKa = 4.62LRR55 pKa = 11.84AKK57 pKa = 10.26VKK59 pKa = 10.73APEE62 pKa = 4.67SYY64 pKa = 10.17TGDD67 pKa = 3.29IPVGNSGLSSIKK79 pKa = 10.33INAEE83 pKa = 3.71KK84 pKa = 10.52IEE86 pKa = 4.21IPNASTMSVVYY97 pKa = 9.47CEE99 pKa = 4.12PLSYY103 pKa = 11.2DD104 pKa = 3.16SDD106 pKa = 4.13YY107 pKa = 11.61KK108 pKa = 10.35KK109 pKa = 9.07TLAEE113 pKa = 4.16SFFDD117 pKa = 3.62KK118 pKa = 11.11SKK120 pKa = 11.28GIYY123 pKa = 9.67KK124 pKa = 9.99YY125 pKa = 10.58DD126 pKa = 3.39YY127 pKa = 8.38EE128 pKa = 4.69KK129 pKa = 10.62PIRR132 pKa = 11.84EE133 pKa = 4.19DD134 pKa = 3.22VQKK137 pKa = 10.99EE138 pKa = 3.89LDD140 pKa = 3.76VYY142 pKa = 11.2QAMLDD147 pKa = 3.81DD148 pKa = 5.28ANAAGDD154 pKa = 3.91TEE156 pKa = 4.14NAQWIGDD163 pKa = 3.94TVNSLKK169 pKa = 10.65SDD171 pKa = 3.67LAKK174 pKa = 10.31ATDD177 pKa = 3.35EE178 pKa = 4.35RR179 pKa = 11.84EE180 pKa = 4.4PVGDD184 pKa = 4.04TYY186 pKa = 11.71DD187 pKa = 3.35SDD189 pKa = 4.86SYY191 pKa = 10.98IGYY194 pKa = 9.8IGDD197 pKa = 3.55QLYY200 pKa = 10.49EE201 pKa = 4.24LDD203 pKa = 4.08MYY205 pKa = 11.36GDD207 pKa = 3.83SSGVNGSASIAPYY220 pKa = 10.01PYY222 pKa = 9.05ITIKK226 pKa = 10.56DD227 pKa = 3.65IRR229 pKa = 11.84PHH231 pKa = 6.56DD232 pKa = 3.86DD233 pKa = 3.81CYY235 pKa = 11.28NVSYY239 pKa = 11.3YY240 pKa = 11.06GDD242 pKa = 4.56DD243 pKa = 3.66YY244 pKa = 11.95NSYY247 pKa = 11.13DD248 pKa = 5.23LEE250 pKa = 4.43NQSDD254 pKa = 3.86MTKK257 pKa = 10.92DD258 pKa = 3.32EE259 pKa = 4.35AVSYY263 pKa = 8.11ATSYY267 pKa = 10.94LADD270 pKa = 4.46LGFKK274 pKa = 10.61DD275 pKa = 5.93AMLKK279 pKa = 10.24SCSPLIWDD287 pKa = 4.0YY288 pKa = 11.82YY289 pKa = 11.14DD290 pKa = 3.47YY291 pKa = 11.68AGNTLAEE298 pKa = 4.47EE299 pKa = 4.39CDD301 pKa = 3.24GWSITFVKK309 pKa = 10.63SVDD312 pKa = 3.41NVPIYY317 pKa = 11.08SPDD320 pKa = 3.18VWNLDD325 pKa = 3.62YY326 pKa = 11.52LDD328 pKa = 4.7TNNVWYY334 pKa = 6.89TTSDD338 pKa = 3.1QTINLSIYY346 pKa = 10.02NGSAFQVNINYY357 pKa = 10.14SLNTIKK363 pKa = 10.58EE364 pKa = 4.15DD365 pKa = 3.61SDD367 pKa = 4.35VEE369 pKa = 4.16LLSWDD374 pKa = 4.68EE375 pKa = 4.06ILKK378 pKa = 10.35SAQEE382 pKa = 4.52KK383 pKa = 10.38IPEE386 pKa = 4.24FYY388 pKa = 10.66KK389 pKa = 10.9DD390 pKa = 3.36NKK392 pKa = 9.7TSYY395 pKa = 11.33LDD397 pKa = 3.93LEE399 pKa = 4.21FDD401 pKa = 3.96KK402 pKa = 11.65VKK404 pKa = 9.77LTYY407 pKa = 10.29FRR409 pKa = 11.84VKK411 pKa = 10.97DD412 pKa = 3.61EE413 pKa = 4.56DD414 pKa = 3.66TTGQYY419 pKa = 11.65KK420 pKa = 10.53LIPVWAFISTSYY432 pKa = 11.19GDD434 pKa = 3.95EE435 pKa = 4.29EE436 pKa = 6.54GDD438 pKa = 3.38TSTEE442 pKa = 3.74EE443 pKa = 3.97DD444 pKa = 3.76TEE446 pKa = 4.19EE447 pKa = 4.16DD448 pKa = 3.89TEE450 pKa = 4.63DD451 pKa = 3.52SAFEE455 pKa = 4.23DD456 pKa = 4.04SYY458 pKa = 10.4PTQLILINAMDD469 pKa = 4.47GSLIDD474 pKa = 3.61VASQLEE480 pKa = 4.48GEE482 pKa = 4.93SNDD485 pKa = 3.59SDD487 pKa = 4.58DD488 pKa = 4.08VTSSTISDD496 pKa = 3.71DD497 pKa = 3.48GSFAVEE503 pKa = 4.18KK504 pKa = 10.52EE505 pKa = 3.77VDD507 pKa = 3.36EE508 pKa = 5.91GDD510 pKa = 3.52IQQ512 pKa = 4.11
Molecular weight: 56.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5JYM3|R5JYM3_9FIRM Radical SAM protein TIGR01212 family OS=Coprococcus sp. CAG:782 OX=1262863 GN=BN781_00598 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.89VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.89VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
659125 |
30 |
2014 |
336.5 |
37.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.285 ± 0.051 | 1.466 ± 0.023 |
6.666 ± 0.054 | 7.114 ± 0.06 |
3.883 ± 0.038 | 7.204 ± 0.048 |
1.565 ± 0.022 | 7.875 ± 0.054 |
7.193 ± 0.047 | 8.069 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.163 ± 0.031 | 4.958 ± 0.04 |
2.947 ± 0.027 | 2.383 ± 0.027 |
4.174 ± 0.044 | 5.884 ± 0.042 |
5.487 ± 0.057 | 7.382 ± 0.045 |
0.723 ± 0.015 | 4.575 ± 0.046 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
