
Synechococcus phage ACG-2014b
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Nereusvirus; Synechococcus virus ACG2014bSyn7803C61
Average proteome isoelectric point is 5.6
Get precalculated fractions of proteins
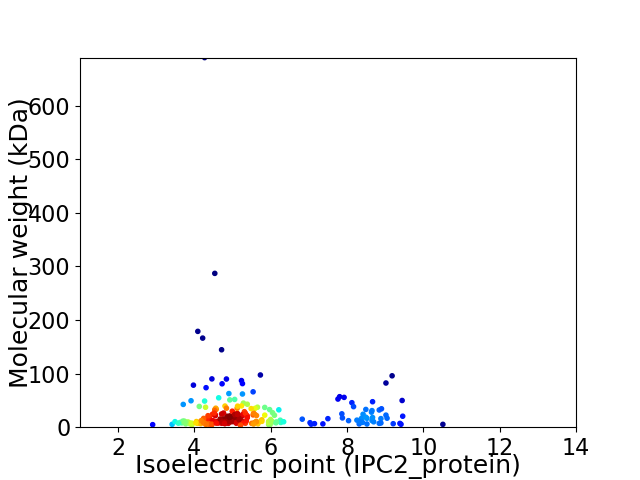
Virtual 2D-PAGE plot for 215 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E3FTG3|A0A0E3FTG3_9CAUD Uncharacterized protein OS=Synechococcus phage ACG-2014b OX=1493508 GN=Syn7803C100_93 PE=4 SV=1
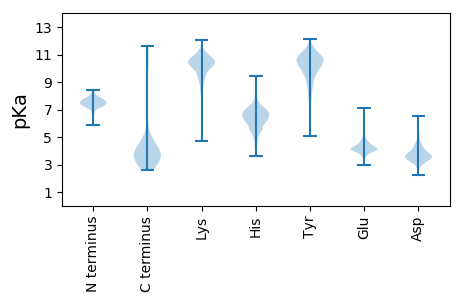
MM1 pKa = 7.77AKK3 pKa = 8.28QTLGLGGAANDD14 pKa = 3.91STGDD18 pKa = 3.61TLRR21 pKa = 11.84VGGDD25 pKa = 3.53KK26 pKa = 11.01INDD29 pKa = 3.71NFDD32 pKa = 4.19EE33 pKa = 4.87IYY35 pKa = 9.5STLGNGSALALTTANAASGHH55 pKa = 4.23VLKK58 pKa = 11.28YY59 pKa = 9.99NGSSFVSADD68 pKa = 3.35FAVLTTSLDD77 pKa = 3.38VDD79 pKa = 3.04IHH81 pKa = 6.46GIISSSDD88 pKa = 3.05RR89 pKa = 11.84NIPITPNGTGDD100 pKa = 3.64VVITAGGTATTFEE113 pKa = 4.68GSDD116 pKa = 3.56GTVDD120 pKa = 4.39FPTSIKK126 pKa = 10.66YY127 pKa = 10.07KK128 pKa = 10.61NEE130 pKa = 3.31YY131 pKa = 8.48TAIGNAPTSATYY143 pKa = 9.62PGYY146 pKa = 10.58FFTVDD151 pKa = 3.65GDD153 pKa = 4.03DD154 pKa = 3.74NPYY157 pKa = 10.99VNMNITAGGVGDD169 pKa = 3.89TQAALLTQYY178 pKa = 11.43SSVDD182 pKa = 3.52DD183 pKa = 4.83LNDD186 pKa = 3.14VDD188 pKa = 4.22TTTNAPANDD197 pKa = 3.68QVLKK201 pKa = 10.25WSASSSKK208 pKa = 9.71WLPQDD213 pKa = 3.88DD214 pKa = 4.25QSGLTNLNLFATITADD230 pKa = 3.5TGSSTADD237 pKa = 3.12SSADD241 pKa = 3.27TLTIAGGSNITTAITGDD258 pKa = 3.47TLTIDD263 pKa = 3.91FSGVVTSTLSGMTDD277 pKa = 2.91TDD279 pKa = 4.07FTGLTQGDD287 pKa = 3.65NLFYY291 pKa = 11.03NGSSWVRR298 pKa = 11.84TASPLTWWEE307 pKa = 4.18LGSSGANDD315 pKa = 3.53YY316 pKa = 8.6TFSGPGFPVTVNDD329 pKa = 3.41PTIYY333 pKa = 9.96VYY335 pKa = 10.69RR336 pKa = 11.84GFTYY340 pKa = 10.82AFDD343 pKa = 3.77NTSNGASHH351 pKa = 7.98PFRR354 pKa = 11.84IQSTTGLSGNPYY366 pKa = 8.16TTGQSGNGQTVLYY379 pKa = 6.82WTVPMDD385 pKa = 4.19SPNTLYY391 pKa = 10.44YY392 pKa = 10.37QCTVHH397 pKa = 7.12AAMNGTIIVVNN408 pKa = 4.33
MM1 pKa = 7.77AKK3 pKa = 8.28QTLGLGGAANDD14 pKa = 3.91STGDD18 pKa = 3.61TLRR21 pKa = 11.84VGGDD25 pKa = 3.53KK26 pKa = 11.01INDD29 pKa = 3.71NFDD32 pKa = 4.19EE33 pKa = 4.87IYY35 pKa = 9.5STLGNGSALALTTANAASGHH55 pKa = 4.23VLKK58 pKa = 11.28YY59 pKa = 9.99NGSSFVSADD68 pKa = 3.35FAVLTTSLDD77 pKa = 3.38VDD79 pKa = 3.04IHH81 pKa = 6.46GIISSSDD88 pKa = 3.05RR89 pKa = 11.84NIPITPNGTGDD100 pKa = 3.64VVITAGGTATTFEE113 pKa = 4.68GSDD116 pKa = 3.56GTVDD120 pKa = 4.39FPTSIKK126 pKa = 10.66YY127 pKa = 10.07KK128 pKa = 10.61NEE130 pKa = 3.31YY131 pKa = 8.48TAIGNAPTSATYY143 pKa = 9.62PGYY146 pKa = 10.58FFTVDD151 pKa = 3.65GDD153 pKa = 4.03DD154 pKa = 3.74NPYY157 pKa = 10.99VNMNITAGGVGDD169 pKa = 3.89TQAALLTQYY178 pKa = 11.43SSVDD182 pKa = 3.52DD183 pKa = 4.83LNDD186 pKa = 3.14VDD188 pKa = 4.22TTTNAPANDD197 pKa = 3.68QVLKK201 pKa = 10.25WSASSSKK208 pKa = 9.71WLPQDD213 pKa = 3.88DD214 pKa = 4.25QSGLTNLNLFATITADD230 pKa = 3.5TGSSTADD237 pKa = 3.12SSADD241 pKa = 3.27TLTIAGGSNITTAITGDD258 pKa = 3.47TLTIDD263 pKa = 3.91FSGVVTSTLSGMTDD277 pKa = 2.91TDD279 pKa = 4.07FTGLTQGDD287 pKa = 3.65NLFYY291 pKa = 11.03NGSSWVRR298 pKa = 11.84TASPLTWWEE307 pKa = 4.18LGSSGANDD315 pKa = 3.53YY316 pKa = 8.6TFSGPGFPVTVNDD329 pKa = 3.41PTIYY333 pKa = 9.96VYY335 pKa = 10.69RR336 pKa = 11.84GFTYY340 pKa = 10.82AFDD343 pKa = 3.77NTSNGASHH351 pKa = 7.98PFRR354 pKa = 11.84IQSTTGLSGNPYY366 pKa = 8.16TTGQSGNGQTVLYY379 pKa = 6.82WTVPMDD385 pKa = 4.19SPNTLYY391 pKa = 10.44YY392 pKa = 10.37QCTVHH397 pKa = 7.12AAMNGTIIVVNN408 pKa = 4.33
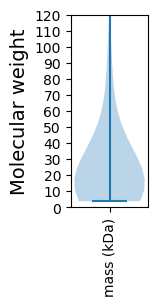
Molecular weight: 42.53 kDa
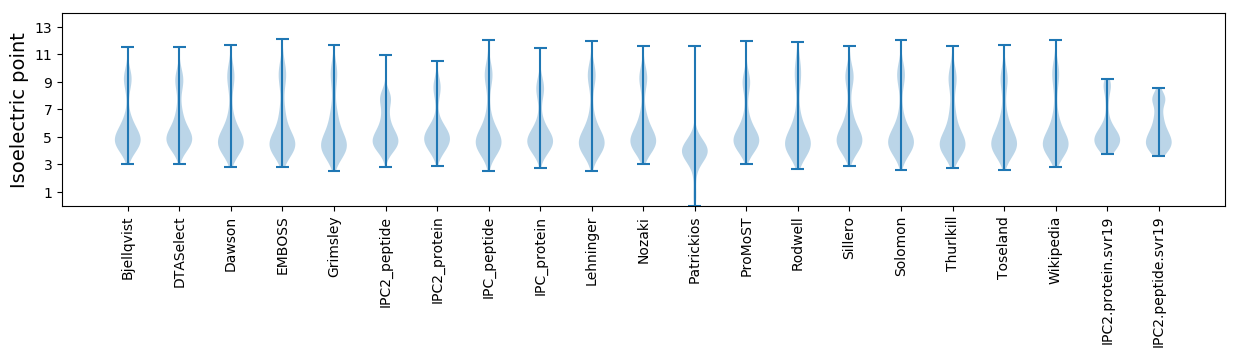
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3F2E4|A0A0E3F2E4_9CAUD Baseplate wedge protein OS=Synechococcus phage ACG-2014b OX=1493508 GN=Syn7803C100_90 PE=4 SV=1
MM1 pKa = 7.51AKK3 pKa = 8.91MRR5 pKa = 11.84KK6 pKa = 8.83SLSGNSMIEE15 pKa = 3.92SSPKK19 pKa = 9.03KK20 pKa = 8.74TRR22 pKa = 11.84QGTGKK27 pKa = 7.38HH28 pKa = 4.73TKK30 pKa = 9.25YY31 pKa = 10.81ASSSRR36 pKa = 11.84NAAKK40 pKa = 10.03KK41 pKa = 10.0RR42 pKa = 11.84YY43 pKa = 9.0RR44 pKa = 11.84GQGKK48 pKa = 9.52
MM1 pKa = 7.51AKK3 pKa = 8.91MRR5 pKa = 11.84KK6 pKa = 8.83SLSGNSMIEE15 pKa = 3.92SSPKK19 pKa = 9.03KK20 pKa = 8.74TRR22 pKa = 11.84QGTGKK27 pKa = 7.38HH28 pKa = 4.73TKK30 pKa = 9.25YY31 pKa = 10.81ASSSRR36 pKa = 11.84NAAKK40 pKa = 10.03KK41 pKa = 10.0RR42 pKa = 11.84YY43 pKa = 9.0RR44 pKa = 11.84GQGKK48 pKa = 9.52
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
54719 |
35 |
6422 |
254.5 |
28.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.818 ± 0.216 | 0.93 ± 0.099 |
6.694 ± 0.131 | 6.256 ± 0.256 |
4.313 ± 0.107 | 7.721 ± 0.325 |
1.413 ± 0.111 | 6.561 ± 0.208 |
5.6 ± 0.386 | 7.23 ± 0.182 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.058 ± 0.221 | 5.903 ± 0.18 |
3.889 ± 0.14 | 3.646 ± 0.099 |
4.008 ± 0.155 | 7.37 ± 0.264 |
7.535 ± 0.426 | 6.773 ± 0.18 |
1.075 ± 0.082 | 4.209 ± 0.131 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
