
Human papillomavirus type 161
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 19
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
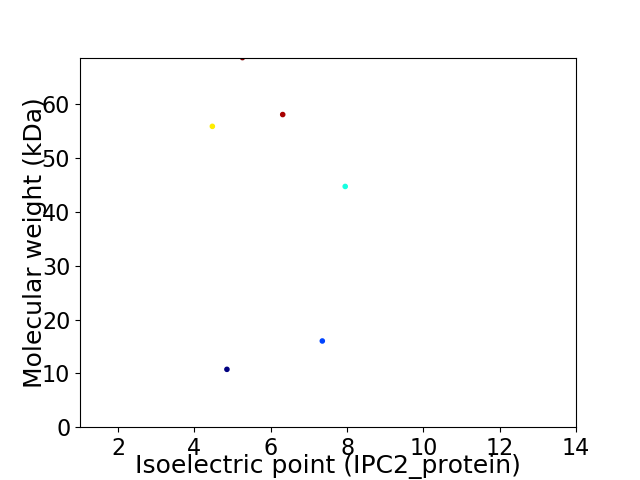
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4MYZ2|K4MYZ2_9PAPI Protein E7 OS=Human papillomavirus type 161 OX=1315264 GN=E7 PE=3 SV=1
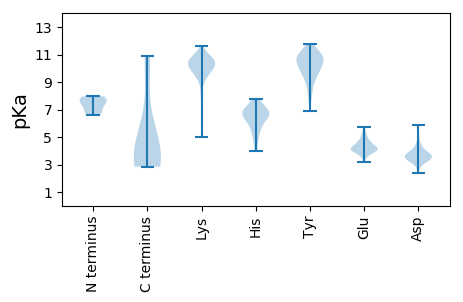
MM1 pKa = 6.86YY2 pKa = 10.0RR3 pKa = 11.84AGRR6 pKa = 11.84RR7 pKa = 11.84TRR9 pKa = 11.84ASAEE13 pKa = 3.99DD14 pKa = 4.32LYY16 pKa = 10.9KK17 pKa = 10.62QCFTGDD23 pKa = 3.78CPPDD27 pKa = 3.41VKK29 pKa = 11.18NKK31 pKa = 10.16IEE33 pKa = 4.24GDD35 pKa = 3.2TWADD39 pKa = 3.91RR40 pKa = 11.84LLKK43 pKa = 10.26WFGSVIYY50 pKa = 10.62LGGLGIGTGRR60 pKa = 11.84GSGGSTGYY68 pKa = 10.46RR69 pKa = 11.84PIGGTTRR76 pKa = 11.84PIPDD80 pKa = 4.88TIPVRR85 pKa = 11.84PAVPVEE91 pKa = 3.99PIGPVEE97 pKa = 5.15IIPIDD102 pKa = 3.97TINPAGPSVIDD113 pKa = 3.64LTNVQIPDD121 pKa = 3.62PSIIDD126 pKa = 3.32IANPTTALGPGGIDD140 pKa = 2.56IVSATDD146 pKa = 4.12PIGDD150 pKa = 3.71LGGVGGNPTVITSVDD165 pKa = 3.36DD166 pKa = 3.32TAAILDD172 pKa = 4.04VQPIPPPKK180 pKa = 10.16RR181 pKa = 11.84FALDD185 pKa = 3.26VNNEE189 pKa = 3.85PRR191 pKa = 11.84SGHH194 pKa = 4.01VTVYY198 pKa = 10.95SSTTHH203 pKa = 7.41PDD205 pKa = 2.87PDD207 pKa = 3.43INIFVSPNYY216 pKa = 9.69QGEE219 pKa = 4.23IVGDD223 pKa = 3.73VEE225 pKa = 5.15EE226 pKa = 4.98IPLTTFSEE234 pKa = 4.35MAEE237 pKa = 4.04FEE239 pKa = 4.06IMEE242 pKa = 5.34PGPQTSTPTQIVEE255 pKa = 4.2RR256 pKa = 11.84LAGNARR262 pKa = 11.84RR263 pKa = 11.84LYY265 pKa = 10.69NRR267 pKa = 11.84LFEE270 pKa = 4.32QVPTRR275 pKa = 11.84NPYY278 pKa = 10.07FLGPVSRR285 pKa = 11.84AVQFEE290 pKa = 4.1YY291 pKa = 11.12SNPVFDD297 pKa = 5.66ADD299 pKa = 3.63VTVTFEE305 pKa = 3.99NDD307 pKa = 3.11LAEE310 pKa = 4.31IAAAPDD316 pKa = 3.23SDD318 pKa = 3.85FRR320 pKa = 11.84DD321 pKa = 3.63VQILHH326 pKa = 6.83RR327 pKa = 11.84PTYY330 pKa = 6.9TTTDD334 pKa = 2.84QGFIRR339 pKa = 11.84VSRR342 pKa = 11.84EE343 pKa = 3.2GARR346 pKa = 11.84ASITTRR352 pKa = 11.84SGLQLGRR359 pKa = 11.84PVHH362 pKa = 6.8FYY364 pKa = 11.03QDD366 pKa = 3.24LSAIEE371 pKa = 4.19SAEE374 pKa = 3.96SFEE377 pKa = 4.05MHH379 pKa = 6.57SLNTTSHH386 pKa = 6.5SSTTVNTLLDD396 pKa = 4.57SITVNPAYY404 pKa = 9.63EE405 pKa = 4.23DD406 pKa = 3.33PFYY409 pKa = 10.66PDD411 pKa = 5.85DD412 pKa = 4.32EE413 pKa = 5.36LLDD416 pKa = 4.4PLDD419 pKa = 3.49EE420 pKa = 4.46TFQRR424 pKa = 11.84GHH426 pKa = 6.19IVLTEE431 pKa = 4.06TNVDD435 pKa = 2.79GDD437 pKa = 4.37IIQFPTTLAEE447 pKa = 4.25SPVKK451 pKa = 10.86VFINDD456 pKa = 3.4YY457 pKa = 9.87GSSITVSYY465 pKa = 8.06PHH467 pKa = 7.02PAEE470 pKa = 4.16TASKK474 pKa = 9.88IIPIPSTDD482 pKa = 3.41NTPLLVLDD490 pKa = 4.93LTSDD494 pKa = 3.99DD495 pKa = 4.46YY496 pKa = 11.62FLHH499 pKa = 6.51PSLRR503 pKa = 11.84KK504 pKa = 9.4RR505 pKa = 11.84KK506 pKa = 9.36RR507 pKa = 11.84KK508 pKa = 9.97RR509 pKa = 11.84SDD511 pKa = 3.0VFF513 pKa = 3.52
MM1 pKa = 6.86YY2 pKa = 10.0RR3 pKa = 11.84AGRR6 pKa = 11.84RR7 pKa = 11.84TRR9 pKa = 11.84ASAEE13 pKa = 3.99DD14 pKa = 4.32LYY16 pKa = 10.9KK17 pKa = 10.62QCFTGDD23 pKa = 3.78CPPDD27 pKa = 3.41VKK29 pKa = 11.18NKK31 pKa = 10.16IEE33 pKa = 4.24GDD35 pKa = 3.2TWADD39 pKa = 3.91RR40 pKa = 11.84LLKK43 pKa = 10.26WFGSVIYY50 pKa = 10.62LGGLGIGTGRR60 pKa = 11.84GSGGSTGYY68 pKa = 10.46RR69 pKa = 11.84PIGGTTRR76 pKa = 11.84PIPDD80 pKa = 4.88TIPVRR85 pKa = 11.84PAVPVEE91 pKa = 3.99PIGPVEE97 pKa = 5.15IIPIDD102 pKa = 3.97TINPAGPSVIDD113 pKa = 3.64LTNVQIPDD121 pKa = 3.62PSIIDD126 pKa = 3.32IANPTTALGPGGIDD140 pKa = 2.56IVSATDD146 pKa = 4.12PIGDD150 pKa = 3.71LGGVGGNPTVITSVDD165 pKa = 3.36DD166 pKa = 3.32TAAILDD172 pKa = 4.04VQPIPPPKK180 pKa = 10.16RR181 pKa = 11.84FALDD185 pKa = 3.26VNNEE189 pKa = 3.85PRR191 pKa = 11.84SGHH194 pKa = 4.01VTVYY198 pKa = 10.95SSTTHH203 pKa = 7.41PDD205 pKa = 2.87PDD207 pKa = 3.43INIFVSPNYY216 pKa = 9.69QGEE219 pKa = 4.23IVGDD223 pKa = 3.73VEE225 pKa = 5.15EE226 pKa = 4.98IPLTTFSEE234 pKa = 4.35MAEE237 pKa = 4.04FEE239 pKa = 4.06IMEE242 pKa = 5.34PGPQTSTPTQIVEE255 pKa = 4.2RR256 pKa = 11.84LAGNARR262 pKa = 11.84RR263 pKa = 11.84LYY265 pKa = 10.69NRR267 pKa = 11.84LFEE270 pKa = 4.32QVPTRR275 pKa = 11.84NPYY278 pKa = 10.07FLGPVSRR285 pKa = 11.84AVQFEE290 pKa = 4.1YY291 pKa = 11.12SNPVFDD297 pKa = 5.66ADD299 pKa = 3.63VTVTFEE305 pKa = 3.99NDD307 pKa = 3.11LAEE310 pKa = 4.31IAAAPDD316 pKa = 3.23SDD318 pKa = 3.85FRR320 pKa = 11.84DD321 pKa = 3.63VQILHH326 pKa = 6.83RR327 pKa = 11.84PTYY330 pKa = 6.9TTTDD334 pKa = 2.84QGFIRR339 pKa = 11.84VSRR342 pKa = 11.84EE343 pKa = 3.2GARR346 pKa = 11.84ASITTRR352 pKa = 11.84SGLQLGRR359 pKa = 11.84PVHH362 pKa = 6.8FYY364 pKa = 11.03QDD366 pKa = 3.24LSAIEE371 pKa = 4.19SAEE374 pKa = 3.96SFEE377 pKa = 4.05MHH379 pKa = 6.57SLNTTSHH386 pKa = 6.5SSTTVNTLLDD396 pKa = 4.57SITVNPAYY404 pKa = 9.63EE405 pKa = 4.23DD406 pKa = 3.33PFYY409 pKa = 10.66PDD411 pKa = 5.85DD412 pKa = 4.32EE413 pKa = 5.36LLDD416 pKa = 4.4PLDD419 pKa = 3.49EE420 pKa = 4.46TFQRR424 pKa = 11.84GHH426 pKa = 6.19IVLTEE431 pKa = 4.06TNVDD435 pKa = 2.79GDD437 pKa = 4.37IIQFPTTLAEE447 pKa = 4.25SPVKK451 pKa = 10.86VFINDD456 pKa = 3.4YY457 pKa = 9.87GSSITVSYY465 pKa = 8.06PHH467 pKa = 7.02PAEE470 pKa = 4.16TASKK474 pKa = 9.88IIPIPSTDD482 pKa = 3.41NTPLLVLDD490 pKa = 4.93LTSDD494 pKa = 3.99DD495 pKa = 4.46YY496 pKa = 11.62FLHH499 pKa = 6.51PSLRR503 pKa = 11.84KK504 pKa = 9.4RR505 pKa = 11.84KK506 pKa = 9.36RR507 pKa = 11.84KK508 pKa = 9.97RR509 pKa = 11.84SDD511 pKa = 3.0VFF513 pKa = 3.52
Molecular weight: 55.89 kDa
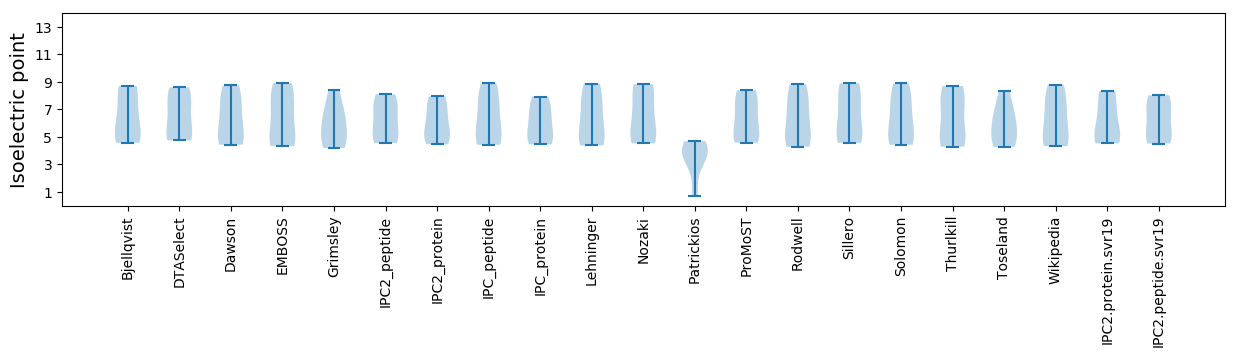
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4MLG5|K4MLG5_9PAPI Major capsid protein L1 OS=Human papillomavirus type 161 OX=1315264 GN=L1 PE=3 SV=1
MM1 pKa = 7.44ACARR5 pKa = 11.84PLRR8 pKa = 11.84LDD10 pKa = 4.02DD11 pKa = 4.05FCAVYY16 pKa = 10.4SVSLFDD22 pKa = 4.7VSLPCIFCKK31 pKa = 10.4FVCDD35 pKa = 4.23LQDD38 pKa = 3.17LAAFHH43 pKa = 6.67IRR45 pKa = 11.84NLCLVWRR52 pKa = 11.84SEE54 pKa = 4.3QAFACCRR61 pKa = 11.84KK62 pKa = 10.0CINLSAKK69 pKa = 10.37YY70 pKa = 9.92EE71 pKa = 3.9YY72 pKa = 10.84DD73 pKa = 3.55NYY75 pKa = 10.71CVCIVNALSIEE86 pKa = 4.02QLLGLSLKK94 pKa = 10.81NIVIRR99 pKa = 11.84CLCCYY104 pKa = 10.12KK105 pKa = 10.81LLDD108 pKa = 3.61IAEE111 pKa = 4.86KK112 pKa = 10.74LDD114 pKa = 3.83CCAGNEE120 pKa = 3.95FFALVRR126 pKa = 11.84GTWRR130 pKa = 11.84GPCRR134 pKa = 11.84NCIRR138 pKa = 11.84KK139 pKa = 9.13RR140 pKa = 3.44
MM1 pKa = 7.44ACARR5 pKa = 11.84PLRR8 pKa = 11.84LDD10 pKa = 4.02DD11 pKa = 4.05FCAVYY16 pKa = 10.4SVSLFDD22 pKa = 4.7VSLPCIFCKK31 pKa = 10.4FVCDD35 pKa = 4.23LQDD38 pKa = 3.17LAAFHH43 pKa = 6.67IRR45 pKa = 11.84NLCLVWRR52 pKa = 11.84SEE54 pKa = 4.3QAFACCRR61 pKa = 11.84KK62 pKa = 10.0CINLSAKK69 pKa = 10.37YY70 pKa = 9.92EE71 pKa = 3.9YY72 pKa = 10.84DD73 pKa = 3.55NYY75 pKa = 10.71CVCIVNALSIEE86 pKa = 4.02QLLGLSLKK94 pKa = 10.81NIVIRR99 pKa = 11.84CLCCYY104 pKa = 10.12KK105 pKa = 10.81LLDD108 pKa = 3.61IAEE111 pKa = 4.86KK112 pKa = 10.74LDD114 pKa = 3.83CCAGNEE120 pKa = 3.95FFALVRR126 pKa = 11.84GTWRR130 pKa = 11.84GPCRR134 pKa = 11.84NCIRR138 pKa = 11.84KK139 pKa = 9.13RR140 pKa = 3.44
Molecular weight: 16.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2253 |
97 |
604 |
375.5 |
42.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.814 ± 0.588 | 2.574 ± 1.13 |
6.968 ± 0.463 | 5.992 ± 0.56 |
4.749 ± 0.323 | 5.548 ± 0.772 |
1.687 ± 0.116 | 5.237 ± 0.778 |
5.237 ± 0.962 | 8.389 ± 0.942 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.687 ± 0.468 | 4.927 ± 0.449 |
6.214 ± 1.079 | 3.551 ± 0.309 |
5.814 ± 0.852 | 7.59 ± 0.635 |
6.436 ± 1.094 | 6.658 ± 0.311 |
1.509 ± 0.346 | 3.418 ± 0.156 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
