
Nocardioides sp. BN130099
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
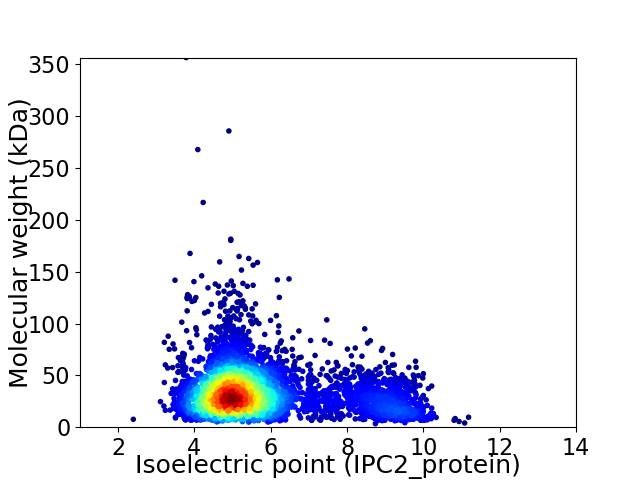
Virtual 2D-PAGE plot for 4452 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B1LKR9|A0A5B1LKR9_9ACTN Ferredoxin OS=Nocardioides sp. BN130099 OX=2607660 GN=F0U44_07175 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.78KK3 pKa = 9.41WIVAIIVVWSLVIVSGIAVAVVLVTGGDD31 pKa = 3.91DD32 pKa = 5.0DD33 pKa = 5.95NDD35 pKa = 3.82AQRR38 pKa = 11.84DD39 pKa = 3.89PSPSPTTPGEE49 pKa = 4.28SSTSPTTPAGPVPAGLEE66 pKa = 4.09SFYY69 pKa = 10.74SQQVAWEE76 pKa = 3.9QCGSDD81 pKa = 2.92KK82 pKa = 11.2CGTLTVPLDD91 pKa = 3.56YY92 pKa = 11.31AEE94 pKa = 4.49PTGEE98 pKa = 4.12TIEE101 pKa = 4.7IALEE105 pKa = 3.75MAPATGEE112 pKa = 4.23RR113 pKa = 11.84IGSLVVNPGGPGAPGTDD130 pKa = 3.15TAKK133 pKa = 10.96DD134 pKa = 3.07ADD136 pKa = 4.32YY137 pKa = 11.45YY138 pKa = 10.49FAPEE142 pKa = 4.18LRR144 pKa = 11.84VAYY147 pKa = 9.71DD148 pKa = 3.06IVGFDD153 pKa = 3.66PRR155 pKa = 11.84GTGDD159 pKa = 4.1SSPIDD164 pKa = 4.37CLPDD168 pKa = 3.92DD169 pKa = 4.77QLDD172 pKa = 3.98EE173 pKa = 4.35YY174 pKa = 11.36LAGDD178 pKa = 4.56PDD180 pKa = 5.39PDD182 pKa = 3.77TKK184 pKa = 11.56AEE186 pKa = 4.13VADD189 pKa = 5.17FNADD193 pKa = 3.47SADD196 pKa = 3.72YY197 pKa = 10.09WSGCAEE203 pKa = 3.93RR204 pKa = 11.84SGDD207 pKa = 3.66LAAHH211 pKa = 6.54VSTIEE216 pKa = 3.71AARR219 pKa = 11.84DD220 pKa = 3.44MDD222 pKa = 4.18VLRR225 pKa = 11.84AALGEE230 pKa = 4.22SQLAYY235 pKa = 10.88LGFSYY240 pKa = 8.75GTRR243 pKa = 11.84LGATYY248 pKa = 10.98ASLFPANVGRR258 pKa = 11.84LVLDD262 pKa = 4.04GAVDD266 pKa = 3.91PSLPTLEE273 pKa = 5.22GSLSQAKK280 pKa = 10.02GFEE283 pKa = 4.19TALRR287 pKa = 11.84SYY289 pKa = 10.49LQNCVDD295 pKa = 5.76LGDD298 pKa = 4.63CFLGDD303 pKa = 4.29SVDD306 pKa = 4.09AGLKK310 pKa = 7.46TVKK313 pKa = 10.46DD314 pKa = 3.82LLADD318 pKa = 3.78IDD320 pKa = 4.11AQPIPTDD327 pKa = 3.9DD328 pKa = 4.25PDD330 pKa = 4.85RR331 pKa = 11.84DD332 pKa = 3.9LTVGRR337 pKa = 11.84AFYY340 pKa = 10.8GVVAPLYY347 pKa = 11.18SKK349 pKa = 11.1DD350 pKa = 2.62NWTYY354 pKa = 11.3LDD356 pKa = 3.47QGLEE360 pKa = 3.9QALDD364 pKa = 3.85GDD366 pKa = 4.35GSTLLFLSDD375 pKa = 5.06FYY377 pKa = 11.0TSRR380 pKa = 11.84NEE382 pKa = 4.09DD383 pKa = 2.54GTYY386 pKa = 10.87ADD388 pKa = 4.47NSVEE392 pKa = 4.3AIGVINCLDD401 pKa = 4.48DD402 pKa = 3.77PWSITSAQVKK412 pKa = 8.13SHH414 pKa = 5.5YY415 pKa = 10.53KK416 pKa = 9.75EE417 pKa = 4.09FEE419 pKa = 3.97KK420 pKa = 10.87ASPTFGDD427 pKa = 3.43VFAWGLVSCNGDD439 pKa = 3.21PFTSTEE445 pKa = 3.75PDD447 pKa = 3.05IPITGAGAAPIVVIGTTRR465 pKa = 11.84DD466 pKa = 3.1PATPYY471 pKa = 11.1AEE473 pKa = 4.08AVAMADD479 pKa = 3.43QLEE482 pKa = 4.46SGVLLSRR489 pKa = 11.84DD490 pKa = 3.62GDD492 pKa = 3.57GHH494 pKa = 4.95TAYY497 pKa = 10.86NKK499 pKa = 10.58GNACIDD505 pKa = 4.22DD506 pKa = 4.07AVHH509 pKa = 7.22AYY511 pKa = 10.57LIDD514 pKa = 3.69GTVPADD520 pKa = 3.5GTEE523 pKa = 4.1CC524 pKa = 4.36
MM1 pKa = 7.44KK2 pKa = 9.78KK3 pKa = 9.41WIVAIIVVWSLVIVSGIAVAVVLVTGGDD31 pKa = 3.91DD32 pKa = 5.0DD33 pKa = 5.95NDD35 pKa = 3.82AQRR38 pKa = 11.84DD39 pKa = 3.89PSPSPTTPGEE49 pKa = 4.28SSTSPTTPAGPVPAGLEE66 pKa = 4.09SFYY69 pKa = 10.74SQQVAWEE76 pKa = 3.9QCGSDD81 pKa = 2.92KK82 pKa = 11.2CGTLTVPLDD91 pKa = 3.56YY92 pKa = 11.31AEE94 pKa = 4.49PTGEE98 pKa = 4.12TIEE101 pKa = 4.7IALEE105 pKa = 3.75MAPATGEE112 pKa = 4.23RR113 pKa = 11.84IGSLVVNPGGPGAPGTDD130 pKa = 3.15TAKK133 pKa = 10.96DD134 pKa = 3.07ADD136 pKa = 4.32YY137 pKa = 11.45YY138 pKa = 10.49FAPEE142 pKa = 4.18LRR144 pKa = 11.84VAYY147 pKa = 9.71DD148 pKa = 3.06IVGFDD153 pKa = 3.66PRR155 pKa = 11.84GTGDD159 pKa = 4.1SSPIDD164 pKa = 4.37CLPDD168 pKa = 3.92DD169 pKa = 4.77QLDD172 pKa = 3.98EE173 pKa = 4.35YY174 pKa = 11.36LAGDD178 pKa = 4.56PDD180 pKa = 5.39PDD182 pKa = 3.77TKK184 pKa = 11.56AEE186 pKa = 4.13VADD189 pKa = 5.17FNADD193 pKa = 3.47SADD196 pKa = 3.72YY197 pKa = 10.09WSGCAEE203 pKa = 3.93RR204 pKa = 11.84SGDD207 pKa = 3.66LAAHH211 pKa = 6.54VSTIEE216 pKa = 3.71AARR219 pKa = 11.84DD220 pKa = 3.44MDD222 pKa = 4.18VLRR225 pKa = 11.84AALGEE230 pKa = 4.22SQLAYY235 pKa = 10.88LGFSYY240 pKa = 8.75GTRR243 pKa = 11.84LGATYY248 pKa = 10.98ASLFPANVGRR258 pKa = 11.84LVLDD262 pKa = 4.04GAVDD266 pKa = 3.91PSLPTLEE273 pKa = 5.22GSLSQAKK280 pKa = 10.02GFEE283 pKa = 4.19TALRR287 pKa = 11.84SYY289 pKa = 10.49LQNCVDD295 pKa = 5.76LGDD298 pKa = 4.63CFLGDD303 pKa = 4.29SVDD306 pKa = 4.09AGLKK310 pKa = 7.46TVKK313 pKa = 10.46DD314 pKa = 3.82LLADD318 pKa = 3.78IDD320 pKa = 4.11AQPIPTDD327 pKa = 3.9DD328 pKa = 4.25PDD330 pKa = 4.85RR331 pKa = 11.84DD332 pKa = 3.9LTVGRR337 pKa = 11.84AFYY340 pKa = 10.8GVVAPLYY347 pKa = 11.18SKK349 pKa = 11.1DD350 pKa = 2.62NWTYY354 pKa = 11.3LDD356 pKa = 3.47QGLEE360 pKa = 3.9QALDD364 pKa = 3.85GDD366 pKa = 4.35GSTLLFLSDD375 pKa = 5.06FYY377 pKa = 11.0TSRR380 pKa = 11.84NEE382 pKa = 4.09DD383 pKa = 2.54GTYY386 pKa = 10.87ADD388 pKa = 4.47NSVEE392 pKa = 4.3AIGVINCLDD401 pKa = 4.48DD402 pKa = 3.77PWSITSAQVKK412 pKa = 8.13SHH414 pKa = 5.5YY415 pKa = 10.53KK416 pKa = 9.75EE417 pKa = 4.09FEE419 pKa = 3.97KK420 pKa = 10.87ASPTFGDD427 pKa = 3.43VFAWGLVSCNGDD439 pKa = 3.21PFTSTEE445 pKa = 3.75PDD447 pKa = 3.05IPITGAGAAPIVVIGTTRR465 pKa = 11.84DD466 pKa = 3.1PATPYY471 pKa = 11.1AEE473 pKa = 4.08AVAMADD479 pKa = 3.43QLEE482 pKa = 4.46SGVLLSRR489 pKa = 11.84DD490 pKa = 3.62GDD492 pKa = 3.57GHH494 pKa = 4.95TAYY497 pKa = 10.86NKK499 pKa = 10.58GNACIDD505 pKa = 4.22DD506 pKa = 4.07AVHH509 pKa = 7.22AYY511 pKa = 10.57LIDD514 pKa = 3.69GTVPADD520 pKa = 3.5GTEE523 pKa = 4.1CC524 pKa = 4.36
Molecular weight: 55.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B1LN51|A0A5B1LN51_9ACTN Uncharacterized protein OS=Nocardioides sp. BN130099 OX=2607660 GN=F0U44_03725 PE=4 SV=1
MM1 pKa = 7.37TATPTPPDD9 pKa = 3.47RR10 pKa = 11.84RR11 pKa = 11.84SSRR14 pKa = 11.84PVRR17 pKa = 11.84TSRR20 pKa = 11.84RR21 pKa = 11.84LPLLRR26 pKa = 11.84PRR28 pKa = 11.84RR29 pKa = 11.84PRR31 pKa = 11.84RR32 pKa = 11.84PPRR35 pKa = 11.84RR36 pKa = 11.84AAPPRR41 pKa = 11.84SRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84WRR47 pKa = 11.84RR48 pKa = 11.84CVRR51 pKa = 11.84PTPSWSRR58 pKa = 11.84PSRR61 pKa = 11.84TPLRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84AIVRR71 pKa = 11.84RR72 pKa = 11.84SPPRR76 pKa = 11.84SRR78 pKa = 11.84RR79 pKa = 3.46
MM1 pKa = 7.37TATPTPPDD9 pKa = 3.47RR10 pKa = 11.84RR11 pKa = 11.84SSRR14 pKa = 11.84PVRR17 pKa = 11.84TSRR20 pKa = 11.84RR21 pKa = 11.84LPLLRR26 pKa = 11.84PRR28 pKa = 11.84RR29 pKa = 11.84PRR31 pKa = 11.84RR32 pKa = 11.84PPRR35 pKa = 11.84RR36 pKa = 11.84AAPPRR41 pKa = 11.84SRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84WRR47 pKa = 11.84RR48 pKa = 11.84CVRR51 pKa = 11.84PTPSWSRR58 pKa = 11.84PSRR61 pKa = 11.84TPLRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84AIVRR71 pKa = 11.84RR72 pKa = 11.84SPPRR76 pKa = 11.84SRR78 pKa = 11.84RR79 pKa = 3.46
Molecular weight: 9.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1471220 |
30 |
3545 |
330.5 |
35.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.679 ± 0.052 | 0.79 ± 0.01 |
6.881 ± 0.037 | 5.607 ± 0.042 |
2.931 ± 0.02 | 9.33 ± 0.051 |
2.116 ± 0.015 | 3.965 ± 0.025 |
2.314 ± 0.027 | 9.767 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.81 ± 0.016 | 2.089 ± 0.028 |
5.522 ± 0.031 | 2.784 ± 0.017 |
7.02 ± 0.047 | 5.397 ± 0.032 |
6.321 ± 0.049 | 9.041 ± 0.037 |
1.495 ± 0.016 | 2.143 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
