
Cervus elaphus papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Xipapillomavirus; Xipapillomavirus 5
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
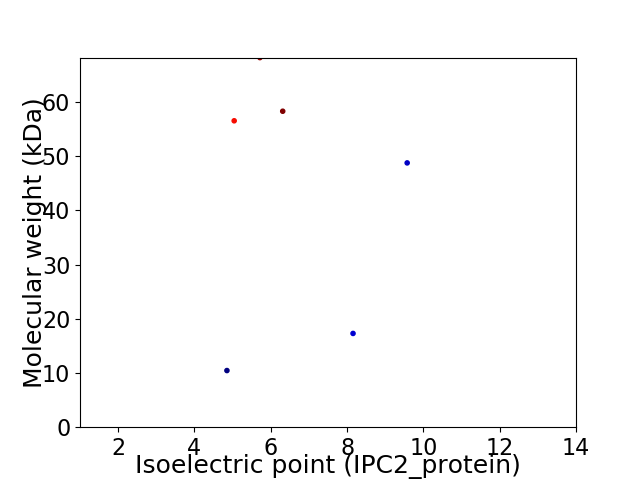
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
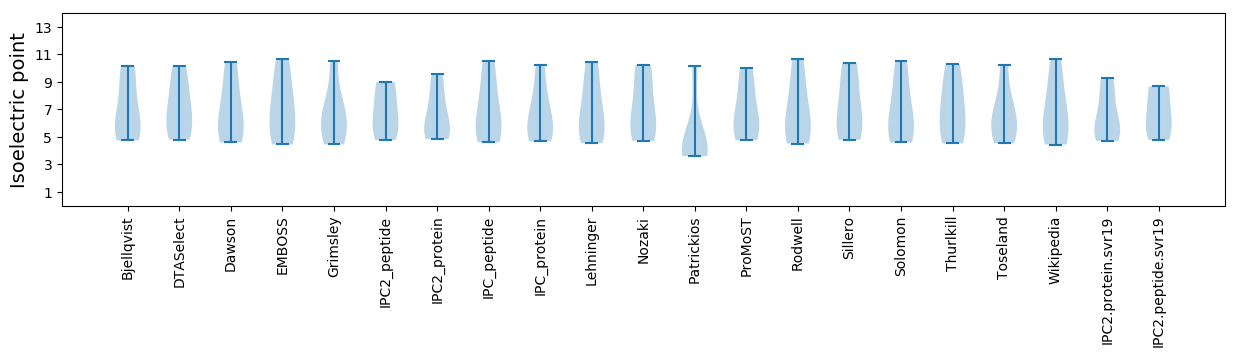
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I9KHZ0|A0A1I9KHZ0_9PAPI Replication protein E1 OS=Cervus elaphus papillomavirus 2 OX=1747359 GN=E1 PE=3 SV=1
MM1 pKa = 7.73RR2 pKa = 11.84GDD4 pKa = 4.04KK5 pKa = 9.39PTISDD10 pKa = 3.28IALDD14 pKa = 4.36LEE16 pKa = 4.68QIVCPVSLEE25 pKa = 4.31CNEE28 pKa = 4.07TMSPEE33 pKa = 3.94EE34 pKa = 4.8AEE36 pKa = 4.34AALQFPYY43 pKa = 10.3RR44 pKa = 11.84VATSCSGCNKK54 pKa = 9.41EE55 pKa = 3.98LHH57 pKa = 6.45LFVAATEE64 pKa = 3.97EE65 pKa = 4.93GIRR68 pKa = 11.84SLEE71 pKa = 3.81VSLIGDD77 pKa = 4.25KK78 pKa = 11.13LSLLCGRR85 pKa = 11.84CARR88 pKa = 11.84VIFSNGQRR96 pKa = 3.41
MM1 pKa = 7.73RR2 pKa = 11.84GDD4 pKa = 4.04KK5 pKa = 9.39PTISDD10 pKa = 3.28IALDD14 pKa = 4.36LEE16 pKa = 4.68QIVCPVSLEE25 pKa = 4.31CNEE28 pKa = 4.07TMSPEE33 pKa = 3.94EE34 pKa = 4.8AEE36 pKa = 4.34AALQFPYY43 pKa = 10.3RR44 pKa = 11.84VATSCSGCNKK54 pKa = 9.41EE55 pKa = 3.98LHH57 pKa = 6.45LFVAATEE64 pKa = 3.97EE65 pKa = 4.93GIRR68 pKa = 11.84SLEE71 pKa = 3.81VSLIGDD77 pKa = 4.25KK78 pKa = 11.13LSLLCGRR85 pKa = 11.84CARR88 pKa = 11.84VIFSNGQRR96 pKa = 3.41
Molecular weight: 10.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I9KHZ8|A0A1I9KHZ8_9PAPI Regulatory protein E2 OS=Cervus elaphus papillomavirus 2 OX=1747359 GN=E2 PE=3 SV=1
MM1 pKa = 7.79EE2 pKa = 4.97NLQARR7 pKa = 11.84FDD9 pKa = 4.01VVQEE13 pKa = 3.98KK14 pKa = 10.65LMEE17 pKa = 4.76IYY19 pKa = 10.46EE20 pKa = 4.48SDD22 pKa = 3.41VQTIDD27 pKa = 3.15KK28 pKa = 10.63QIEE31 pKa = 3.74HH32 pKa = 6.42WQTVRR37 pKa = 11.84LEE39 pKa = 4.02QVLLFVARR47 pKa = 11.84KK48 pKa = 9.85KK49 pKa = 11.0GITRR53 pKa = 11.84MGLQVVPPTAVSEE66 pKa = 4.19ARR68 pKa = 11.84AKK70 pKa = 10.55NAIEE74 pKa = 3.89MVLVLEE80 pKa = 4.28RR81 pKa = 11.84LKK83 pKa = 11.19DD84 pKa = 3.52SAYY87 pKa = 9.45GTEE90 pKa = 4.48KK91 pKa = 8.78WTLTDD96 pKa = 4.21SSRR99 pKa = 11.84EE100 pKa = 4.09TYY102 pKa = 9.83KK103 pKa = 10.85AAPEE107 pKa = 3.96DD108 pKa = 4.04TFKK111 pKa = 10.97KK112 pKa = 10.01RR113 pKa = 11.84PASVTVIYY121 pKa = 10.94DD122 pKa = 3.51GDD124 pKa = 4.1AYY126 pKa = 11.64NSMIYY131 pKa = 9.5TMWRR135 pKa = 11.84DD136 pKa = 2.91IYY138 pKa = 11.51YY139 pKa = 10.51EE140 pKa = 5.5DD141 pKa = 3.34DD142 pKa = 3.98TGWHH146 pKa = 6.18KK147 pKa = 11.07VPSQVDD153 pKa = 3.77YY154 pKa = 11.51YY155 pKa = 11.05GIYY158 pKa = 10.09YY159 pKa = 9.81VDD161 pKa = 4.0HH162 pKa = 6.82EE163 pKa = 4.86GEE165 pKa = 3.79KK166 pKa = 10.2KK167 pKa = 10.82YY168 pKa = 11.19YY169 pKa = 10.57VLFEE173 pKa = 4.74KK174 pKa = 10.92DD175 pKa = 2.72ADD177 pKa = 4.0LYY179 pKa = 11.26SRR181 pKa = 11.84NGQWEE186 pKa = 4.25VQFEE190 pKa = 4.17NMQFSAPVTSSTTGSSRR207 pKa = 11.84HH208 pKa = 5.32GPEE211 pKa = 3.7TASTRR216 pKa = 11.84EE217 pKa = 4.05DD218 pKa = 3.61PQASSAGPPAARR230 pKa = 11.84PRR232 pKa = 11.84QPASPTRR239 pKa = 11.84RR240 pKa = 11.84RR241 pKa = 11.84HH242 pKa = 5.13RR243 pKa = 11.84APLVSSLLSHH253 pKa = 6.4RR254 pKa = 11.84RR255 pKa = 11.84SRR257 pKa = 11.84SRR259 pKa = 11.84SGSSRR264 pKa = 11.84SSRR267 pKa = 11.84SSRR270 pKa = 11.84SSRR273 pKa = 11.84SRR275 pKa = 11.84SPAKK279 pKa = 9.81RR280 pKa = 11.84PRR282 pKa = 11.84RR283 pKa = 11.84RR284 pKa = 11.84GRR286 pKa = 11.84RR287 pKa = 11.84GRR289 pKa = 11.84RR290 pKa = 11.84VARR293 pKa = 11.84SGSRR297 pKa = 11.84GSSSDD302 pKa = 3.09SSRR305 pKa = 11.84TEE307 pKa = 3.69PGGQSPPSPEE317 pKa = 3.86QVGEE321 pKa = 4.08RR322 pKa = 11.84SRR324 pKa = 11.84LPPRR328 pKa = 11.84EE329 pKa = 3.8ASSRR333 pKa = 11.84LARR336 pKa = 11.84LISEE340 pKa = 4.38ARR342 pKa = 11.84DD343 pKa = 3.54PPVLLVKK350 pKa = 10.59GSPNVLRR357 pKa = 11.84CWRR360 pKa = 11.84RR361 pKa = 11.84RR362 pKa = 11.84CRR364 pKa = 11.84LRR366 pKa = 11.84YY367 pKa = 8.78SAYY370 pKa = 8.34FQSMTTTFTWISGHH384 pKa = 4.82STDD387 pKa = 4.15RR388 pKa = 11.84LGGNRR393 pKa = 11.84MLVGFADD400 pKa = 4.7TIQRR404 pKa = 11.84NEE406 pKa = 4.06FLNKK410 pKa = 9.02VKK412 pKa = 10.01MPKK415 pKa = 9.85RR416 pKa = 11.84VTWAYY421 pKa = 10.37GALDD425 pKa = 3.61TFF427 pKa = 4.81
MM1 pKa = 7.79EE2 pKa = 4.97NLQARR7 pKa = 11.84FDD9 pKa = 4.01VVQEE13 pKa = 3.98KK14 pKa = 10.65LMEE17 pKa = 4.76IYY19 pKa = 10.46EE20 pKa = 4.48SDD22 pKa = 3.41VQTIDD27 pKa = 3.15KK28 pKa = 10.63QIEE31 pKa = 3.74HH32 pKa = 6.42WQTVRR37 pKa = 11.84LEE39 pKa = 4.02QVLLFVARR47 pKa = 11.84KK48 pKa = 9.85KK49 pKa = 11.0GITRR53 pKa = 11.84MGLQVVPPTAVSEE66 pKa = 4.19ARR68 pKa = 11.84AKK70 pKa = 10.55NAIEE74 pKa = 3.89MVLVLEE80 pKa = 4.28RR81 pKa = 11.84LKK83 pKa = 11.19DD84 pKa = 3.52SAYY87 pKa = 9.45GTEE90 pKa = 4.48KK91 pKa = 8.78WTLTDD96 pKa = 4.21SSRR99 pKa = 11.84EE100 pKa = 4.09TYY102 pKa = 9.83KK103 pKa = 10.85AAPEE107 pKa = 3.96DD108 pKa = 4.04TFKK111 pKa = 10.97KK112 pKa = 10.01RR113 pKa = 11.84PASVTVIYY121 pKa = 10.94DD122 pKa = 3.51GDD124 pKa = 4.1AYY126 pKa = 11.64NSMIYY131 pKa = 9.5TMWRR135 pKa = 11.84DD136 pKa = 2.91IYY138 pKa = 11.51YY139 pKa = 10.51EE140 pKa = 5.5DD141 pKa = 3.34DD142 pKa = 3.98TGWHH146 pKa = 6.18KK147 pKa = 11.07VPSQVDD153 pKa = 3.77YY154 pKa = 11.51YY155 pKa = 11.05GIYY158 pKa = 10.09YY159 pKa = 9.81VDD161 pKa = 4.0HH162 pKa = 6.82EE163 pKa = 4.86GEE165 pKa = 3.79KK166 pKa = 10.2KK167 pKa = 10.82YY168 pKa = 11.19YY169 pKa = 10.57VLFEE173 pKa = 4.74KK174 pKa = 10.92DD175 pKa = 2.72ADD177 pKa = 4.0LYY179 pKa = 11.26SRR181 pKa = 11.84NGQWEE186 pKa = 4.25VQFEE190 pKa = 4.17NMQFSAPVTSSTTGSSRR207 pKa = 11.84HH208 pKa = 5.32GPEE211 pKa = 3.7TASTRR216 pKa = 11.84EE217 pKa = 4.05DD218 pKa = 3.61PQASSAGPPAARR230 pKa = 11.84PRR232 pKa = 11.84QPASPTRR239 pKa = 11.84RR240 pKa = 11.84RR241 pKa = 11.84HH242 pKa = 5.13RR243 pKa = 11.84APLVSSLLSHH253 pKa = 6.4RR254 pKa = 11.84RR255 pKa = 11.84SRR257 pKa = 11.84SRR259 pKa = 11.84SGSSRR264 pKa = 11.84SSRR267 pKa = 11.84SSRR270 pKa = 11.84SSRR273 pKa = 11.84SRR275 pKa = 11.84SPAKK279 pKa = 9.81RR280 pKa = 11.84PRR282 pKa = 11.84RR283 pKa = 11.84RR284 pKa = 11.84GRR286 pKa = 11.84RR287 pKa = 11.84GRR289 pKa = 11.84RR290 pKa = 11.84VARR293 pKa = 11.84SGSRR297 pKa = 11.84GSSSDD302 pKa = 3.09SSRR305 pKa = 11.84TEE307 pKa = 3.69PGGQSPPSPEE317 pKa = 3.86QVGEE321 pKa = 4.08RR322 pKa = 11.84SRR324 pKa = 11.84LPPRR328 pKa = 11.84EE329 pKa = 3.8ASSRR333 pKa = 11.84LARR336 pKa = 11.84LISEE340 pKa = 4.38ARR342 pKa = 11.84DD343 pKa = 3.54PPVLLVKK350 pKa = 10.59GSPNVLRR357 pKa = 11.84CWRR360 pKa = 11.84RR361 pKa = 11.84RR362 pKa = 11.84CRR364 pKa = 11.84LRR366 pKa = 11.84YY367 pKa = 8.78SAYY370 pKa = 8.34FQSMTTTFTWISGHH384 pKa = 4.82STDD387 pKa = 4.15RR388 pKa = 11.84LGGNRR393 pKa = 11.84MLVGFADD400 pKa = 4.7TIQRR404 pKa = 11.84NEE406 pKa = 4.06FLNKK410 pKa = 9.02VKK412 pKa = 10.01MPKK415 pKa = 9.85RR416 pKa = 11.84VTWAYY421 pKa = 10.37GALDD425 pKa = 3.61TFF427 pKa = 4.81
Molecular weight: 48.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2297 |
96 |
601 |
382.8 |
43.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.834 ± 0.637 | 2.264 ± 0.81 |
5.921 ± 0.338 | 6.182 ± 0.401 |
4.615 ± 0.482 | 6.487 ± 0.692 |
1.785 ± 0.093 | 4.441 ± 0.675 |
5.05 ± 0.612 | 8.489 ± 0.807 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.48 ± 0.227 | 4.179 ± 0.614 |
6.313 ± 0.847 | 4.136 ± 0.331 |
7.357 ± 1.216 | 8.098 ± 0.872 |
6.356 ± 0.556 | 6.051 ± 0.284 |
1.524 ± 0.311 | 3.439 ± 0.306 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
