
Aspergillus fumigatus partitivirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 8.18
Get precalculated fractions of proteins
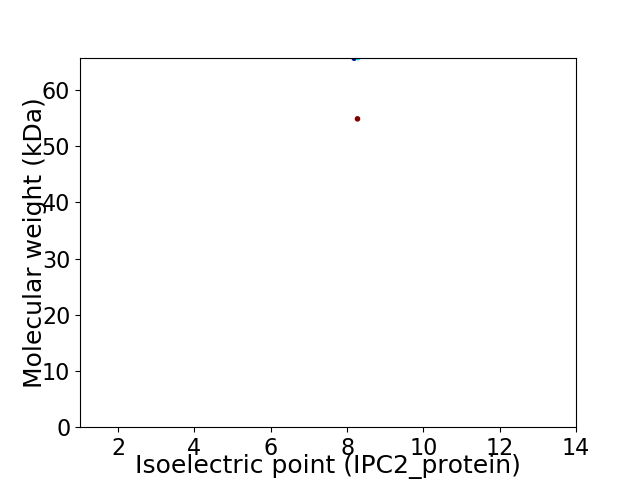
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A344X3Q1|A0A344X3Q1_9VIRU Coat protein OS=Aspergillus fumigatus partitivirus 2 OX=2250452 PE=4 SV=1
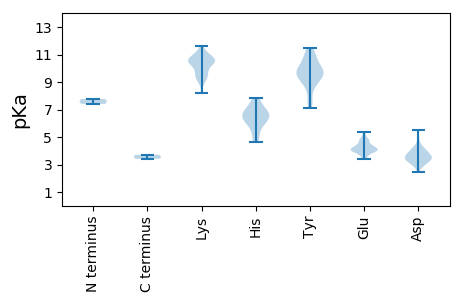
MM1 pKa = 7.76LLTEE5 pKa = 4.83IPNVYY10 pKa = 9.55QRR12 pKa = 11.84AVALEE17 pKa = 3.5RR18 pKa = 11.84RR19 pKa = 11.84IRR21 pKa = 11.84LRR23 pKa = 11.84EE24 pKa = 3.99LYY26 pKa = 9.99PEE28 pKa = 4.45GVVPYY33 pKa = 9.87LAKK36 pKa = 10.25QEE38 pKa = 4.0YY39 pKa = 9.73DD40 pKa = 3.32GAIAGPHH47 pKa = 6.09GPHH50 pKa = 6.66GSVAPSVTTTFQAKK64 pKa = 10.11LPLDD68 pKa = 3.5QYY70 pKa = 11.09PDD72 pKa = 2.91RR73 pKa = 11.84KK74 pKa = 9.18YY75 pKa = 9.2TLEE78 pKa = 4.19CKK80 pKa = 9.11EE81 pKa = 4.02TMIYY85 pKa = 9.18LGKK88 pKa = 9.38MDD90 pKa = 5.39GYY92 pKa = 9.36PFSSSIPKK100 pKa = 9.62FDD102 pKa = 2.79PWFRR106 pKa = 11.84KK107 pKa = 8.56VLKK110 pKa = 10.45DD111 pKa = 3.41KK112 pKa = 11.22DD113 pKa = 3.5PAIVKK118 pKa = 10.11DD119 pKa = 4.17LEE121 pKa = 4.31DD122 pKa = 3.03TFMRR126 pKa = 11.84DD127 pKa = 2.47PCTPDD132 pKa = 3.26RR133 pKa = 11.84VMKK136 pKa = 10.68HH137 pKa = 5.03MSLFDD142 pKa = 3.87RR143 pKa = 11.84TWKK146 pKa = 10.46RR147 pKa = 11.84MPTGRR152 pKa = 11.84LMSRR156 pKa = 11.84AKK158 pKa = 10.72DD159 pKa = 3.15IVAKK163 pKa = 9.7MFEE166 pKa = 4.78PIGKK170 pKa = 9.2VEE172 pKa = 4.81PIDD175 pKa = 3.94FNFAGWHH182 pKa = 6.16KK183 pKa = 10.4ILPNLDD189 pKa = 3.54MTSSPGLPLRR199 pKa = 11.84RR200 pKa = 11.84EE201 pKa = 3.99YY202 pKa = 9.28QTQGEE207 pKa = 4.78CLGHH211 pKa = 7.8IYY213 pKa = 10.92DD214 pKa = 3.46KK215 pKa = 10.74TKK217 pKa = 10.79RR218 pKa = 11.84LNHH221 pKa = 6.11FAKK224 pKa = 9.98FLHH227 pKa = 6.44PAAVRR232 pKa = 11.84APPCMIGVRR241 pKa = 11.84PGLIRR246 pKa = 11.84RR247 pKa = 11.84DD248 pKa = 3.54EE249 pKa = 4.2YY250 pKa = 11.46NEE252 pKa = 3.93KK253 pKa = 10.5VKK255 pKa = 10.98ARR257 pKa = 11.84GVWAYY262 pKa = 8.57PAEE265 pKa = 4.33VKK267 pKa = 10.51VMEE270 pKa = 4.24MRR272 pKa = 11.84FVQPLIEE279 pKa = 4.34RR280 pKa = 11.84MSLNFMKK287 pKa = 10.19IPYY290 pKa = 9.09PVGRR294 pKa = 11.84NMTKK298 pKa = 10.48ALPMLIDD305 pKa = 4.84HH306 pKa = 6.8MLHH309 pKa = 6.61DD310 pKa = 4.34KK311 pKa = 11.08KK312 pKa = 10.95KK313 pKa = 10.71GFVTDD318 pKa = 3.47VSNLDD323 pKa = 3.44ASVGPDD329 pKa = 3.44YY330 pKa = 11.06IDD332 pKa = 3.08WAFSLMKK339 pKa = 9.47TWFNFGITQSSEE351 pKa = 3.55TRR353 pKa = 11.84NANVFDD359 pKa = 3.88FLHH362 pKa = 6.77YY363 pKa = 9.89YY364 pKa = 10.23FKK366 pKa = 10.31RR367 pKa = 11.84TPILLPSGQLVKK379 pKa = 10.69KK380 pKa = 10.69AGGVPSGSGFTQLVDD395 pKa = 3.52TLVTILATVYY405 pKa = 9.84TLLRR409 pKa = 11.84MGWDD413 pKa = 3.28EE414 pKa = 4.02DD415 pKa = 4.11QIIKK419 pKa = 10.81GYY421 pKa = 7.41MQVVGDD427 pKa = 4.4DD428 pKa = 3.71MAVSVPRR435 pKa = 11.84DD436 pKa = 3.38FDD438 pKa = 3.81PEE440 pKa = 4.04EE441 pKa = 3.86FVYY444 pKa = 10.86LMGSLGFTINLKK456 pKa = 10.55KK457 pKa = 11.25VMFSDD462 pKa = 4.23KK463 pKa = 10.96GIEE466 pKa = 4.11LKK468 pKa = 10.83FLGYY472 pKa = 9.85SKK474 pKa = 11.18YY475 pKa = 10.23GGSVYY480 pKa = 10.49RR481 pKa = 11.84PLDD484 pKa = 3.69EE485 pKa = 5.16LLQTAFFPEE494 pKa = 4.37KK495 pKa = 10.36FVGNPEE501 pKa = 3.98RR502 pKa = 11.84SRR504 pKa = 11.84MRR506 pKa = 11.84IAGQTLASGMTNAFFSKK523 pKa = 9.6VNYY526 pKa = 9.05WMEE529 pKa = 4.19EE530 pKa = 4.11LVSLATEE537 pKa = 4.63LDD539 pKa = 3.55PDD541 pKa = 3.94EE542 pKa = 5.23VFIPQRR548 pKa = 11.84RR549 pKa = 11.84WLRR552 pKa = 11.84IVLGLDD558 pKa = 3.22EE559 pKa = 4.94MPSTTLAFDD568 pKa = 3.74MFPLVV573 pKa = 3.42
MM1 pKa = 7.76LLTEE5 pKa = 4.83IPNVYY10 pKa = 9.55QRR12 pKa = 11.84AVALEE17 pKa = 3.5RR18 pKa = 11.84RR19 pKa = 11.84IRR21 pKa = 11.84LRR23 pKa = 11.84EE24 pKa = 3.99LYY26 pKa = 9.99PEE28 pKa = 4.45GVVPYY33 pKa = 9.87LAKK36 pKa = 10.25QEE38 pKa = 4.0YY39 pKa = 9.73DD40 pKa = 3.32GAIAGPHH47 pKa = 6.09GPHH50 pKa = 6.66GSVAPSVTTTFQAKK64 pKa = 10.11LPLDD68 pKa = 3.5QYY70 pKa = 11.09PDD72 pKa = 2.91RR73 pKa = 11.84KK74 pKa = 9.18YY75 pKa = 9.2TLEE78 pKa = 4.19CKK80 pKa = 9.11EE81 pKa = 4.02TMIYY85 pKa = 9.18LGKK88 pKa = 9.38MDD90 pKa = 5.39GYY92 pKa = 9.36PFSSSIPKK100 pKa = 9.62FDD102 pKa = 2.79PWFRR106 pKa = 11.84KK107 pKa = 8.56VLKK110 pKa = 10.45DD111 pKa = 3.41KK112 pKa = 11.22DD113 pKa = 3.5PAIVKK118 pKa = 10.11DD119 pKa = 4.17LEE121 pKa = 4.31DD122 pKa = 3.03TFMRR126 pKa = 11.84DD127 pKa = 2.47PCTPDD132 pKa = 3.26RR133 pKa = 11.84VMKK136 pKa = 10.68HH137 pKa = 5.03MSLFDD142 pKa = 3.87RR143 pKa = 11.84TWKK146 pKa = 10.46RR147 pKa = 11.84MPTGRR152 pKa = 11.84LMSRR156 pKa = 11.84AKK158 pKa = 10.72DD159 pKa = 3.15IVAKK163 pKa = 9.7MFEE166 pKa = 4.78PIGKK170 pKa = 9.2VEE172 pKa = 4.81PIDD175 pKa = 3.94FNFAGWHH182 pKa = 6.16KK183 pKa = 10.4ILPNLDD189 pKa = 3.54MTSSPGLPLRR199 pKa = 11.84RR200 pKa = 11.84EE201 pKa = 3.99YY202 pKa = 9.28QTQGEE207 pKa = 4.78CLGHH211 pKa = 7.8IYY213 pKa = 10.92DD214 pKa = 3.46KK215 pKa = 10.74TKK217 pKa = 10.79RR218 pKa = 11.84LNHH221 pKa = 6.11FAKK224 pKa = 9.98FLHH227 pKa = 6.44PAAVRR232 pKa = 11.84APPCMIGVRR241 pKa = 11.84PGLIRR246 pKa = 11.84RR247 pKa = 11.84DD248 pKa = 3.54EE249 pKa = 4.2YY250 pKa = 11.46NEE252 pKa = 3.93KK253 pKa = 10.5VKK255 pKa = 10.98ARR257 pKa = 11.84GVWAYY262 pKa = 8.57PAEE265 pKa = 4.33VKK267 pKa = 10.51VMEE270 pKa = 4.24MRR272 pKa = 11.84FVQPLIEE279 pKa = 4.34RR280 pKa = 11.84MSLNFMKK287 pKa = 10.19IPYY290 pKa = 9.09PVGRR294 pKa = 11.84NMTKK298 pKa = 10.48ALPMLIDD305 pKa = 4.84HH306 pKa = 6.8MLHH309 pKa = 6.61DD310 pKa = 4.34KK311 pKa = 11.08KK312 pKa = 10.95KK313 pKa = 10.71GFVTDD318 pKa = 3.47VSNLDD323 pKa = 3.44ASVGPDD329 pKa = 3.44YY330 pKa = 11.06IDD332 pKa = 3.08WAFSLMKK339 pKa = 9.47TWFNFGITQSSEE351 pKa = 3.55TRR353 pKa = 11.84NANVFDD359 pKa = 3.88FLHH362 pKa = 6.77YY363 pKa = 9.89YY364 pKa = 10.23FKK366 pKa = 10.31RR367 pKa = 11.84TPILLPSGQLVKK379 pKa = 10.69KK380 pKa = 10.69AGGVPSGSGFTQLVDD395 pKa = 3.52TLVTILATVYY405 pKa = 9.84TLLRR409 pKa = 11.84MGWDD413 pKa = 3.28EE414 pKa = 4.02DD415 pKa = 4.11QIIKK419 pKa = 10.81GYY421 pKa = 7.41MQVVGDD427 pKa = 4.4DD428 pKa = 3.71MAVSVPRR435 pKa = 11.84DD436 pKa = 3.38FDD438 pKa = 3.81PEE440 pKa = 4.04EE441 pKa = 3.86FVYY444 pKa = 10.86LMGSLGFTINLKK456 pKa = 10.55KK457 pKa = 11.25VMFSDD462 pKa = 4.23KK463 pKa = 10.96GIEE466 pKa = 4.11LKK468 pKa = 10.83FLGYY472 pKa = 9.85SKK474 pKa = 11.18YY475 pKa = 10.23GGSVYY480 pKa = 10.49RR481 pKa = 11.84PLDD484 pKa = 3.69EE485 pKa = 5.16LLQTAFFPEE494 pKa = 4.37KK495 pKa = 10.36FVGNPEE501 pKa = 3.98RR502 pKa = 11.84SRR504 pKa = 11.84MRR506 pKa = 11.84IAGQTLASGMTNAFFSKK523 pKa = 9.6VNYY526 pKa = 9.05WMEE529 pKa = 4.19EE530 pKa = 4.11LVSLATEE537 pKa = 4.63LDD539 pKa = 3.55PDD541 pKa = 3.94EE542 pKa = 5.23VFIPQRR548 pKa = 11.84RR549 pKa = 11.84WLRR552 pKa = 11.84IVLGLDD558 pKa = 3.22EE559 pKa = 4.94MPSTTLAFDD568 pKa = 3.74MFPLVV573 pKa = 3.42
Molecular weight: 65.64 kDa
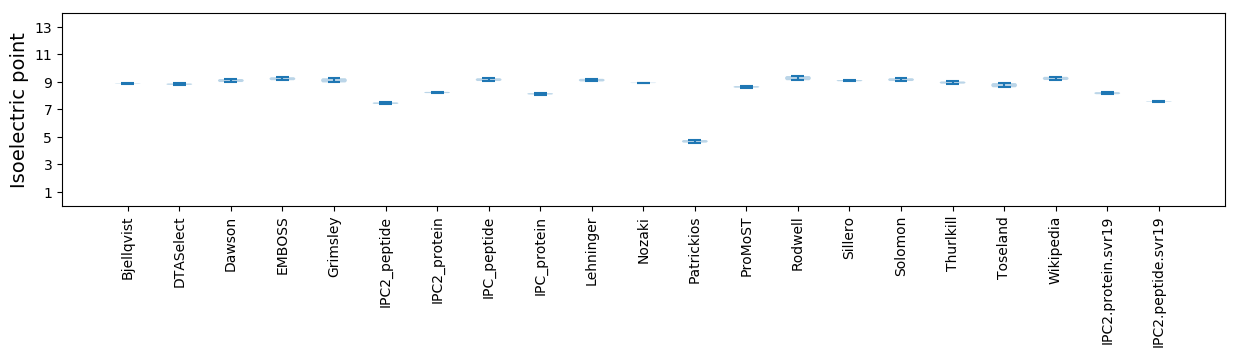
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A344X3Q1|A0A344X3Q1_9VIRU Coat protein OS=Aspergillus fumigatus partitivirus 2 OX=2250452 PE=4 SV=1
MM1 pKa = 7.41PRR3 pKa = 11.84KK4 pKa = 8.91DD5 pKa = 3.39TAEE8 pKa = 4.31KK9 pKa = 11.11DD10 pKa = 3.57PVTKK14 pKa = 10.52DD15 pKa = 3.12DD16 pKa = 3.87TKK18 pKa = 10.59GQKK21 pKa = 9.57DD22 pKa = 3.38AAADD26 pKa = 3.68STDD29 pKa = 3.17KK30 pKa = 11.59GKK32 pKa = 11.17GNKK35 pKa = 9.34PKK37 pKa = 8.99TQKK40 pKa = 10.56QKK42 pKa = 9.72TEE44 pKa = 4.01SSASKK49 pKa = 8.23YY50 pKa = 9.09TPGFAEE56 pKa = 4.84GNSVLQGYY64 pKa = 8.47NLSVSGRR71 pKa = 11.84MEE73 pKa = 5.38RR74 pKa = 11.84ILSPFKK80 pKa = 10.52GDD82 pKa = 3.25KK83 pKa = 10.25FFHH86 pKa = 6.32FVSDD90 pKa = 3.07AWSEE94 pKa = 4.09FTRR97 pKa = 11.84LKK99 pKa = 10.27PHH101 pKa = 4.62ITEE104 pKa = 3.99RR105 pKa = 11.84FSYY108 pKa = 11.28AEE110 pKa = 4.02FRR112 pKa = 11.84HH113 pKa = 5.59MSALQLYY120 pKa = 9.33NRR122 pKa = 11.84LEE124 pKa = 4.02AVKK127 pKa = 10.21FDD129 pKa = 3.95ALGVKK134 pKa = 9.86QPAPTRR140 pKa = 11.84IPLPRR145 pKa = 11.84DD146 pKa = 3.13TRR148 pKa = 11.84VFQPIWSVLANIGVVEE164 pKa = 4.7DD165 pKa = 3.84PDD167 pKa = 3.64LRR169 pKa = 11.84AIYY172 pKa = 9.86IPDD175 pKa = 4.78GIVPKK180 pKa = 10.73SKK182 pKa = 10.58DD183 pKa = 3.47LSHH186 pKa = 7.52PDD188 pKa = 4.12DD189 pKa = 4.08IEE191 pKa = 4.52NLLSCTLYY199 pKa = 10.75DD200 pKa = 4.19WEE202 pKa = 4.73SSWKK206 pKa = 9.48EE207 pKa = 3.57VVKK210 pKa = 10.85ARR212 pKa = 11.84EE213 pKa = 3.82ARR215 pKa = 11.84KK216 pKa = 9.51QYY218 pKa = 7.72QQRR221 pKa = 11.84TGLDD225 pKa = 2.94ATTDD229 pKa = 3.65SNEE232 pKa = 4.12TTVQLSKK239 pKa = 11.41EE240 pKa = 3.94EE241 pKa = 4.73LIAKK245 pKa = 9.25IQFFRR250 pKa = 11.84KK251 pKa = 9.2EE252 pKa = 3.37ITRR255 pKa = 11.84AEE257 pKa = 4.09EE258 pKa = 4.13RR259 pKa = 11.84EE260 pKa = 3.91KK261 pKa = 11.06SPEE264 pKa = 3.81YY265 pKa = 10.55EE266 pKa = 3.92LVKK269 pKa = 10.81GVLRR273 pKa = 11.84KK274 pKa = 9.7IKK276 pKa = 10.51AVTAPSSPSSSRR288 pKa = 11.84SGKK291 pKa = 10.43DD292 pKa = 2.77NGTEE296 pKa = 3.62ATSPKK301 pKa = 10.02KK302 pKa = 10.41EE303 pKa = 4.16YY304 pKa = 10.12EE305 pKa = 3.98QEE307 pKa = 4.43TYY309 pKa = 7.56WTVSGAKK316 pKa = 9.14NQLEE320 pKa = 4.81AYY322 pKa = 7.1MQQAKK327 pKa = 10.0KK328 pKa = 10.59LKK330 pKa = 9.88SQMIAPKK337 pKa = 10.18FDD339 pKa = 3.38VTHH342 pKa = 6.89KK343 pKa = 10.03IEE345 pKa = 4.23SYY347 pKa = 10.17RR348 pKa = 11.84ISDD351 pKa = 3.69GTITTSPGSYY361 pKa = 9.68GEE363 pKa = 3.96WMRR366 pKa = 11.84WDD368 pKa = 3.35PQLYY372 pKa = 8.74IDD374 pKa = 4.34YY375 pKa = 9.47EE376 pKa = 4.39NFVTEE381 pKa = 4.29VTPMALFSLSMPAEE395 pKa = 4.33SKK397 pKa = 9.45GTYY400 pKa = 9.76AWILPVEE407 pKa = 4.22KK408 pKa = 10.62RR409 pKa = 11.84EE410 pKa = 4.45DD411 pKa = 3.89DD412 pKa = 4.07DD413 pKa = 5.53SSVSARR419 pKa = 11.84MPRR422 pKa = 11.84ASIPTATWVLALLLQSSTLPLHH444 pKa = 7.0RR445 pKa = 11.84RR446 pKa = 11.84STWYY450 pKa = 9.69TEE452 pKa = 3.67TDD454 pKa = 2.88RR455 pKa = 11.84LQNVLGVRR463 pKa = 11.84RR464 pKa = 11.84RR465 pKa = 11.84YY466 pKa = 9.52IKK468 pKa = 10.63AAIKK472 pKa = 10.62DD473 pKa = 3.73PTAVEE478 pKa = 4.13QYY480 pKa = 9.63GTII483 pKa = 3.71
MM1 pKa = 7.41PRR3 pKa = 11.84KK4 pKa = 8.91DD5 pKa = 3.39TAEE8 pKa = 4.31KK9 pKa = 11.11DD10 pKa = 3.57PVTKK14 pKa = 10.52DD15 pKa = 3.12DD16 pKa = 3.87TKK18 pKa = 10.59GQKK21 pKa = 9.57DD22 pKa = 3.38AAADD26 pKa = 3.68STDD29 pKa = 3.17KK30 pKa = 11.59GKK32 pKa = 11.17GNKK35 pKa = 9.34PKK37 pKa = 8.99TQKK40 pKa = 10.56QKK42 pKa = 9.72TEE44 pKa = 4.01SSASKK49 pKa = 8.23YY50 pKa = 9.09TPGFAEE56 pKa = 4.84GNSVLQGYY64 pKa = 8.47NLSVSGRR71 pKa = 11.84MEE73 pKa = 5.38RR74 pKa = 11.84ILSPFKK80 pKa = 10.52GDD82 pKa = 3.25KK83 pKa = 10.25FFHH86 pKa = 6.32FVSDD90 pKa = 3.07AWSEE94 pKa = 4.09FTRR97 pKa = 11.84LKK99 pKa = 10.27PHH101 pKa = 4.62ITEE104 pKa = 3.99RR105 pKa = 11.84FSYY108 pKa = 11.28AEE110 pKa = 4.02FRR112 pKa = 11.84HH113 pKa = 5.59MSALQLYY120 pKa = 9.33NRR122 pKa = 11.84LEE124 pKa = 4.02AVKK127 pKa = 10.21FDD129 pKa = 3.95ALGVKK134 pKa = 9.86QPAPTRR140 pKa = 11.84IPLPRR145 pKa = 11.84DD146 pKa = 3.13TRR148 pKa = 11.84VFQPIWSVLANIGVVEE164 pKa = 4.7DD165 pKa = 3.84PDD167 pKa = 3.64LRR169 pKa = 11.84AIYY172 pKa = 9.86IPDD175 pKa = 4.78GIVPKK180 pKa = 10.73SKK182 pKa = 10.58DD183 pKa = 3.47LSHH186 pKa = 7.52PDD188 pKa = 4.12DD189 pKa = 4.08IEE191 pKa = 4.52NLLSCTLYY199 pKa = 10.75DD200 pKa = 4.19WEE202 pKa = 4.73SSWKK206 pKa = 9.48EE207 pKa = 3.57VVKK210 pKa = 10.85ARR212 pKa = 11.84EE213 pKa = 3.82ARR215 pKa = 11.84KK216 pKa = 9.51QYY218 pKa = 7.72QQRR221 pKa = 11.84TGLDD225 pKa = 2.94ATTDD229 pKa = 3.65SNEE232 pKa = 4.12TTVQLSKK239 pKa = 11.41EE240 pKa = 3.94EE241 pKa = 4.73LIAKK245 pKa = 9.25IQFFRR250 pKa = 11.84KK251 pKa = 9.2EE252 pKa = 3.37ITRR255 pKa = 11.84AEE257 pKa = 4.09EE258 pKa = 4.13RR259 pKa = 11.84EE260 pKa = 3.91KK261 pKa = 11.06SPEE264 pKa = 3.81YY265 pKa = 10.55EE266 pKa = 3.92LVKK269 pKa = 10.81GVLRR273 pKa = 11.84KK274 pKa = 9.7IKK276 pKa = 10.51AVTAPSSPSSSRR288 pKa = 11.84SGKK291 pKa = 10.43DD292 pKa = 2.77NGTEE296 pKa = 3.62ATSPKK301 pKa = 10.02KK302 pKa = 10.41EE303 pKa = 4.16YY304 pKa = 10.12EE305 pKa = 3.98QEE307 pKa = 4.43TYY309 pKa = 7.56WTVSGAKK316 pKa = 9.14NQLEE320 pKa = 4.81AYY322 pKa = 7.1MQQAKK327 pKa = 10.0KK328 pKa = 10.59LKK330 pKa = 9.88SQMIAPKK337 pKa = 10.18FDD339 pKa = 3.38VTHH342 pKa = 6.89KK343 pKa = 10.03IEE345 pKa = 4.23SYY347 pKa = 10.17RR348 pKa = 11.84ISDD351 pKa = 3.69GTITTSPGSYY361 pKa = 9.68GEE363 pKa = 3.96WMRR366 pKa = 11.84WDD368 pKa = 3.35PQLYY372 pKa = 8.74IDD374 pKa = 4.34YY375 pKa = 9.47EE376 pKa = 4.39NFVTEE381 pKa = 4.29VTPMALFSLSMPAEE395 pKa = 4.33SKK397 pKa = 9.45GTYY400 pKa = 9.76AWILPVEE407 pKa = 4.22KK408 pKa = 10.62RR409 pKa = 11.84EE410 pKa = 4.45DD411 pKa = 3.89DD412 pKa = 4.07DD413 pKa = 5.53SSVSARR419 pKa = 11.84MPRR422 pKa = 11.84ASIPTATWVLALLLQSSTLPLHH444 pKa = 7.0RR445 pKa = 11.84RR446 pKa = 11.84STWYY450 pKa = 9.69TEE452 pKa = 3.67TDD454 pKa = 2.88RR455 pKa = 11.84LQNVLGVRR463 pKa = 11.84RR464 pKa = 11.84RR465 pKa = 11.84YY466 pKa = 9.52IKK468 pKa = 10.63AAIKK472 pKa = 10.62DD473 pKa = 3.73PTAVEE478 pKa = 4.13QYY480 pKa = 9.63GTII483 pKa = 3.71
Molecular weight: 54.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1056 |
483 |
573 |
528.0 |
60.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.345 ± 0.767 | 0.473 ± 0.184 |
6.345 ± 0.092 | 6.25 ± 0.832 |
4.64 ± 1.061 | 5.682 ± 0.779 |
1.515 ± 0.189 | 4.735 ± 0.019 |
7.67 ± 0.709 | 8.428 ± 0.96 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.598 ± 1.2 | 2.462 ± 0.128 |
6.723 ± 0.497 | 3.409 ± 0.649 |
6.155 ± 0.039 | 6.818 ± 1.584 |
6.534 ± 0.922 | 6.439 ± 0.587 |
1.799 ± 0.187 | 3.977 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
