
Salarchaeum sp. JOR-1
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halobacteriaceae; Salarchaeum; unclassified Salarchaeum
Average proteome isoelectric point is 4.88
Get precalculated fractions of proteins
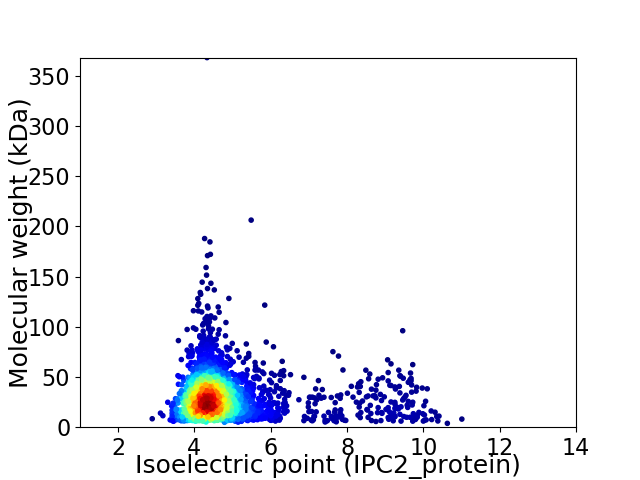
Virtual 2D-PAGE plot for 2527 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A518S9F0|A0A518S9F0_9EURY Plasmid stabilization protein OS=Salarchaeum sp. JOR-1 OX=2599399 GN=FQU85_03685 PE=4 SV=1
MM1 pKa = 7.72NDD3 pKa = 3.55DD4 pKa = 3.55TNDD7 pKa = 3.46RR8 pKa = 11.84QLYY11 pKa = 7.99NLSRR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 10.02VLGGLGAVGLASAGAGLGTSAYY39 pKa = 8.79FTDD42 pKa = 3.96TEE44 pKa = 4.72SFTGNTLAAGEE55 pKa = 4.67LDD57 pKa = 4.13LKK59 pKa = 11.04LDD61 pKa = 3.8YY62 pKa = 10.94KK63 pKa = 10.22STYY66 pKa = 8.55TGGPGRR72 pKa = 11.84LEE74 pKa = 5.57DD75 pKa = 3.98IQAMGYY81 pKa = 9.76PDD83 pKa = 5.61AEE85 pKa = 4.32DD86 pKa = 4.69LGDD89 pKa = 3.77GRR91 pKa = 11.84YY92 pKa = 9.82LLAQTPRR99 pKa = 11.84PGDD102 pKa = 3.37GMGWEE107 pKa = 4.77DD108 pKa = 4.34YY109 pKa = 11.08LPEE112 pKa = 5.14FDD114 pKa = 5.22FCAPEE119 pKa = 3.86NDD121 pKa = 3.47QYY123 pKa = 11.84LVNGDD128 pKa = 4.18GISIFHH134 pKa = 6.97LEE136 pKa = 4.11DD137 pKa = 3.43VKK139 pKa = 11.26PGDD142 pKa = 3.82SGEE145 pKa = 4.24MTVSIHH151 pKa = 6.67ICDD154 pKa = 3.52NPAWLEE160 pKa = 3.9MGGEE164 pKa = 4.02LSEE167 pKa = 4.6NADD170 pKa = 3.38NTMSEE175 pKa = 4.54PEE177 pKa = 4.28SEE179 pKa = 4.28VDD181 pKa = 3.47STPDD185 pKa = 3.15AGEE188 pKa = 3.89LADD191 pKa = 5.0AIEE194 pKa = 4.15VTVWYY199 pKa = 10.19DD200 pKa = 3.41EE201 pKa = 5.36DD202 pKa = 4.4CDD204 pKa = 4.11NVYY207 pKa = 10.8EE208 pKa = 4.09PTGTGEE214 pKa = 4.02QQEE217 pKa = 4.62LEE219 pKa = 4.31VALVSDD225 pKa = 4.32VSGSMSQEE233 pKa = 3.55IGDD236 pKa = 4.38LKK238 pKa = 10.83SAAKK242 pKa = 10.58GFVDD246 pKa = 3.95NLSSPDD252 pKa = 3.19EE253 pKa = 4.02AAAISFSDD261 pKa = 4.64GSSLDD266 pKa = 3.51QQLTTNYY273 pKa = 7.86QTVKK277 pKa = 10.79DD278 pKa = 5.02AIDD281 pKa = 3.49QYY283 pKa = 10.94TSGGGTDD290 pKa = 3.48MDD292 pKa = 3.91QGISTAEE299 pKa = 3.89TEE301 pKa = 4.91LISGANATSGASKK314 pKa = 11.08VMIVLTDD321 pKa = 3.95GQPSSQQAATTAAMNAKK338 pKa = 9.38QAGIRR343 pKa = 11.84IFTIALGSGAATSYY357 pKa = 11.09LEE359 pKa = 4.22NNIASSPGDD368 pKa = 3.45AYY370 pKa = 11.07VAPDD374 pKa = 4.33PSDD377 pKa = 4.09LDD379 pKa = 3.71TVYY382 pKa = 11.06SEE384 pKa = 3.93IAQVVLAGEE393 pKa = 3.96QLIAQGSLADD403 pKa = 4.18VMALLAGGVEE413 pKa = 4.22LDD415 pKa = 3.55GDD417 pKa = 3.75RR418 pKa = 11.84TEE420 pKa = 5.17EE421 pKa = 4.09GRR423 pKa = 11.84QPYY426 pKa = 9.28PGNTTQCIGFEE437 pKa = 4.26WVLPTDD443 pKa = 3.73VEE445 pKa = 4.65NEE447 pKa = 4.1VQTDD451 pKa = 3.65SVAFNVDD458 pKa = 3.85FTATQYY464 pKa = 10.93RR465 pKa = 11.84HH466 pKa = 6.06NPQTTTTAPQQ476 pKa = 3.04
MM1 pKa = 7.72NDD3 pKa = 3.55DD4 pKa = 3.55TNDD7 pKa = 3.46RR8 pKa = 11.84QLYY11 pKa = 7.99NLSRR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 10.02VLGGLGAVGLASAGAGLGTSAYY39 pKa = 8.79FTDD42 pKa = 3.96TEE44 pKa = 4.72SFTGNTLAAGEE55 pKa = 4.67LDD57 pKa = 4.13LKK59 pKa = 11.04LDD61 pKa = 3.8YY62 pKa = 10.94KK63 pKa = 10.22STYY66 pKa = 8.55TGGPGRR72 pKa = 11.84LEE74 pKa = 5.57DD75 pKa = 3.98IQAMGYY81 pKa = 9.76PDD83 pKa = 5.61AEE85 pKa = 4.32DD86 pKa = 4.69LGDD89 pKa = 3.77GRR91 pKa = 11.84YY92 pKa = 9.82LLAQTPRR99 pKa = 11.84PGDD102 pKa = 3.37GMGWEE107 pKa = 4.77DD108 pKa = 4.34YY109 pKa = 11.08LPEE112 pKa = 5.14FDD114 pKa = 5.22FCAPEE119 pKa = 3.86NDD121 pKa = 3.47QYY123 pKa = 11.84LVNGDD128 pKa = 4.18GISIFHH134 pKa = 6.97LEE136 pKa = 4.11DD137 pKa = 3.43VKK139 pKa = 11.26PGDD142 pKa = 3.82SGEE145 pKa = 4.24MTVSIHH151 pKa = 6.67ICDD154 pKa = 3.52NPAWLEE160 pKa = 3.9MGGEE164 pKa = 4.02LSEE167 pKa = 4.6NADD170 pKa = 3.38NTMSEE175 pKa = 4.54PEE177 pKa = 4.28SEE179 pKa = 4.28VDD181 pKa = 3.47STPDD185 pKa = 3.15AGEE188 pKa = 3.89LADD191 pKa = 5.0AIEE194 pKa = 4.15VTVWYY199 pKa = 10.19DD200 pKa = 3.41EE201 pKa = 5.36DD202 pKa = 4.4CDD204 pKa = 4.11NVYY207 pKa = 10.8EE208 pKa = 4.09PTGTGEE214 pKa = 4.02QQEE217 pKa = 4.62LEE219 pKa = 4.31VALVSDD225 pKa = 4.32VSGSMSQEE233 pKa = 3.55IGDD236 pKa = 4.38LKK238 pKa = 10.83SAAKK242 pKa = 10.58GFVDD246 pKa = 3.95NLSSPDD252 pKa = 3.19EE253 pKa = 4.02AAAISFSDD261 pKa = 4.64GSSLDD266 pKa = 3.51QQLTTNYY273 pKa = 7.86QTVKK277 pKa = 10.79DD278 pKa = 5.02AIDD281 pKa = 3.49QYY283 pKa = 10.94TSGGGTDD290 pKa = 3.48MDD292 pKa = 3.91QGISTAEE299 pKa = 3.89TEE301 pKa = 4.91LISGANATSGASKK314 pKa = 11.08VMIVLTDD321 pKa = 3.95GQPSSQQAATTAAMNAKK338 pKa = 9.38QAGIRR343 pKa = 11.84IFTIALGSGAATSYY357 pKa = 11.09LEE359 pKa = 4.22NNIASSPGDD368 pKa = 3.45AYY370 pKa = 11.07VAPDD374 pKa = 4.33PSDD377 pKa = 4.09LDD379 pKa = 3.71TVYY382 pKa = 11.06SEE384 pKa = 3.93IAQVVLAGEE393 pKa = 3.96QLIAQGSLADD403 pKa = 4.18VMALLAGGVEE413 pKa = 4.22LDD415 pKa = 3.55GDD417 pKa = 3.75RR418 pKa = 11.84TEE420 pKa = 5.17EE421 pKa = 4.09GRR423 pKa = 11.84QPYY426 pKa = 9.28PGNTTQCIGFEE437 pKa = 4.26WVLPTDD443 pKa = 3.73VEE445 pKa = 4.65NEE447 pKa = 4.1VQTDD451 pKa = 3.65SVAFNVDD458 pKa = 3.85FTATQYY464 pKa = 10.93RR465 pKa = 11.84HH466 pKa = 6.06NPQTTTTAPQQ476 pKa = 3.04
Molecular weight: 50.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A518S9T5|A0A518S9T5_9EURY Polymer-forming cytoskeletal protein OS=Salarchaeum sp. JOR-1 OX=2599399 GN=FQU85_04265 PE=4 SV=1
MM1 pKa = 7.42TLLPSRR7 pKa = 11.84STLRR11 pKa = 11.84AAAFVVLGTAGFAVSFTMTGQWIRR35 pKa = 11.84WHH37 pKa = 5.75MMGSRR42 pKa = 11.84HH43 pKa = 6.42SFSLQNGVLAALFTGLFALLALVGGIAIARR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 3.59
MM1 pKa = 7.42TLLPSRR7 pKa = 11.84STLRR11 pKa = 11.84AAAFVVLGTAGFAVSFTMTGQWIRR35 pKa = 11.84WHH37 pKa = 5.75MMGSRR42 pKa = 11.84HH43 pKa = 6.42SFSLQNGVLAALFTGLFALLALVGGIAIARR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 3.59
Molecular weight: 8.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
744264 |
37 |
3266 |
294.5 |
31.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.476 ± 0.086 | 0.661 ± 0.015 |
8.6 ± 0.078 | 8.101 ± 0.089 |
3.393 ± 0.035 | 8.266 ± 0.045 |
2.083 ± 0.026 | 3.585 ± 0.04 |
1.921 ± 0.03 | 9.056 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.664 ± 0.019 | 2.457 ± 0.032 |
4.552 ± 0.032 | 2.461 ± 0.032 |
6.519 ± 0.05 | 5.367 ± 0.033 |
6.441 ± 0.06 | 9.42 ± 0.057 |
1.116 ± 0.018 | 2.862 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
