
Cyclovirus TN18
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; Human associated cyclovirus 4
Average proteome isoelectric point is 8.61
Get precalculated fractions of proteins
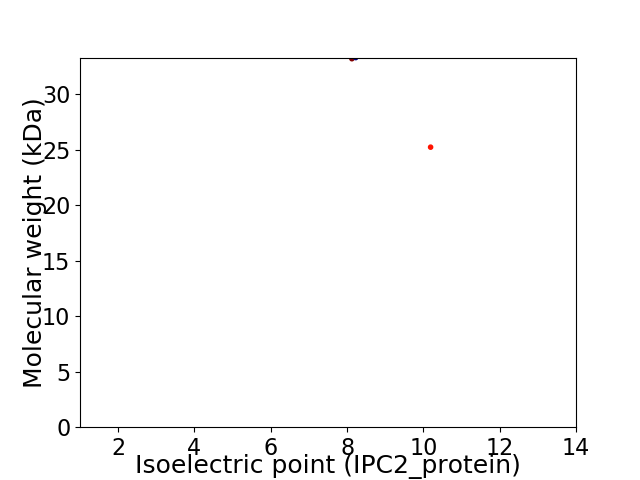
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D4N3R5|D4N3R5_9CIRC Capsid protein OS=Cyclovirus TN18 OX=742925 GN=cap PE=4 SV=1
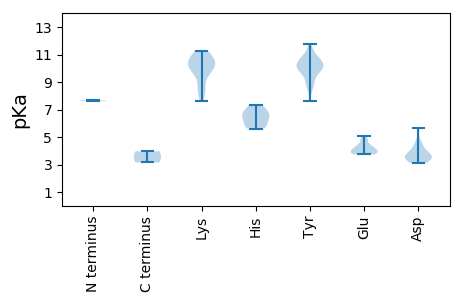
MM1 pKa = 7.61AGSSSKK7 pKa = 8.08QTNSTLRR14 pKa = 11.84RR15 pKa = 11.84FCWTLNNYY23 pKa = 7.61TEE25 pKa = 4.37EE26 pKa = 5.04DD27 pKa = 3.87VTTLQKK33 pKa = 10.99DD34 pKa = 3.35LAEE37 pKa = 4.3LCKK40 pKa = 10.04FAIFGRR46 pKa = 11.84EE47 pKa = 3.79TCPNTGTKK55 pKa = 9.83HH56 pKa = 5.86LQGFCNLQRR65 pKa = 11.84PKK67 pKa = 10.38RR68 pKa = 11.84FSSIRR73 pKa = 11.84KK74 pKa = 8.26LFKK77 pKa = 10.37EE78 pKa = 3.8RR79 pKa = 11.84AHH81 pKa = 6.43IEE83 pKa = 3.96KK84 pKa = 10.77AKK86 pKa = 11.0GSDD89 pKa = 3.81LDD91 pKa = 3.76NKK93 pKa = 8.82TYY95 pKa = 10.26CSKK98 pKa = 10.93SGEE101 pKa = 4.13VWMHH105 pKa = 6.59GEE107 pKa = 4.11PCSQGARR114 pKa = 11.84NDD116 pKa = 3.59LQEE119 pKa = 4.17VVSVIEE125 pKa = 4.21GGEE128 pKa = 4.01RR129 pKa = 11.84NIKK132 pKa = 9.99AVALQFPTTYY142 pKa = 9.86IKK144 pKa = 10.48YY145 pKa = 10.24FKK147 pKa = 10.71GIEE150 pKa = 3.74QYY152 pKa = 10.81IRR154 pKa = 11.84ICHH157 pKa = 5.98SSAEE161 pKa = 4.27RR162 pKa = 11.84DD163 pKa = 3.29FATQVSFFWGPTGSGKK179 pKa = 8.81SRR181 pKa = 11.84RR182 pKa = 11.84AYY184 pKa = 10.1EE185 pKa = 3.79EE186 pKa = 3.89AKK188 pKa = 10.04ATGEE192 pKa = 4.01PIYY195 pKa = 10.23YY196 pKa = 9.93KK197 pKa = 10.49PRR199 pKa = 11.84GEE201 pKa = 3.89WWDD204 pKa = 4.58GYY206 pKa = 10.5CGHH209 pKa = 7.34ANVIIDD215 pKa = 5.69DD216 pKa = 4.49FYY218 pKa = 11.78GWLKK222 pKa = 10.72YY223 pKa = 10.67DD224 pKa = 4.31EE225 pKa = 4.82LLKK228 pKa = 10.7ICDD231 pKa = 3.7RR232 pKa = 11.84YY233 pKa = 10.06PYY235 pKa = 9.86RR236 pKa = 11.84VPVKK240 pKa = 10.11GGYY243 pKa = 9.57EE244 pKa = 3.82NFVTKK249 pKa = 10.4RR250 pKa = 11.84IWITSEE256 pKa = 3.85KK257 pKa = 9.9PLEE260 pKa = 3.98QIYY263 pKa = 10.37RR264 pKa = 11.84FIGYY268 pKa = 8.81DD269 pKa = 3.11CSSIRR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84LNTEE280 pKa = 4.07LYY282 pKa = 9.96IGYY285 pKa = 8.96QQ286 pKa = 3.15
MM1 pKa = 7.61AGSSSKK7 pKa = 8.08QTNSTLRR14 pKa = 11.84RR15 pKa = 11.84FCWTLNNYY23 pKa = 7.61TEE25 pKa = 4.37EE26 pKa = 5.04DD27 pKa = 3.87VTTLQKK33 pKa = 10.99DD34 pKa = 3.35LAEE37 pKa = 4.3LCKK40 pKa = 10.04FAIFGRR46 pKa = 11.84EE47 pKa = 3.79TCPNTGTKK55 pKa = 9.83HH56 pKa = 5.86LQGFCNLQRR65 pKa = 11.84PKK67 pKa = 10.38RR68 pKa = 11.84FSSIRR73 pKa = 11.84KK74 pKa = 8.26LFKK77 pKa = 10.37EE78 pKa = 3.8RR79 pKa = 11.84AHH81 pKa = 6.43IEE83 pKa = 3.96KK84 pKa = 10.77AKK86 pKa = 11.0GSDD89 pKa = 3.81LDD91 pKa = 3.76NKK93 pKa = 8.82TYY95 pKa = 10.26CSKK98 pKa = 10.93SGEE101 pKa = 4.13VWMHH105 pKa = 6.59GEE107 pKa = 4.11PCSQGARR114 pKa = 11.84NDD116 pKa = 3.59LQEE119 pKa = 4.17VVSVIEE125 pKa = 4.21GGEE128 pKa = 4.01RR129 pKa = 11.84NIKK132 pKa = 9.99AVALQFPTTYY142 pKa = 9.86IKK144 pKa = 10.48YY145 pKa = 10.24FKK147 pKa = 10.71GIEE150 pKa = 3.74QYY152 pKa = 10.81IRR154 pKa = 11.84ICHH157 pKa = 5.98SSAEE161 pKa = 4.27RR162 pKa = 11.84DD163 pKa = 3.29FATQVSFFWGPTGSGKK179 pKa = 8.81SRR181 pKa = 11.84RR182 pKa = 11.84AYY184 pKa = 10.1EE185 pKa = 3.79EE186 pKa = 3.89AKK188 pKa = 10.04ATGEE192 pKa = 4.01PIYY195 pKa = 10.23YY196 pKa = 9.93KK197 pKa = 10.49PRR199 pKa = 11.84GEE201 pKa = 3.89WWDD204 pKa = 4.58GYY206 pKa = 10.5CGHH209 pKa = 7.34ANVIIDD215 pKa = 5.69DD216 pKa = 4.49FYY218 pKa = 11.78GWLKK222 pKa = 10.72YY223 pKa = 10.67DD224 pKa = 4.31EE225 pKa = 4.82LLKK228 pKa = 10.7ICDD231 pKa = 3.7RR232 pKa = 11.84YY233 pKa = 10.06PYY235 pKa = 9.86RR236 pKa = 11.84VPVKK240 pKa = 10.11GGYY243 pKa = 9.57EE244 pKa = 3.82NFVTKK249 pKa = 10.4RR250 pKa = 11.84IWITSEE256 pKa = 3.85KK257 pKa = 9.9PLEE260 pKa = 3.98QIYY263 pKa = 10.37RR264 pKa = 11.84FIGYY268 pKa = 8.81DD269 pKa = 3.11CSSIRR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84LNTEE280 pKa = 4.07LYY282 pKa = 9.96IGYY285 pKa = 8.96QQ286 pKa = 3.15
Molecular weight: 33.2 kDa
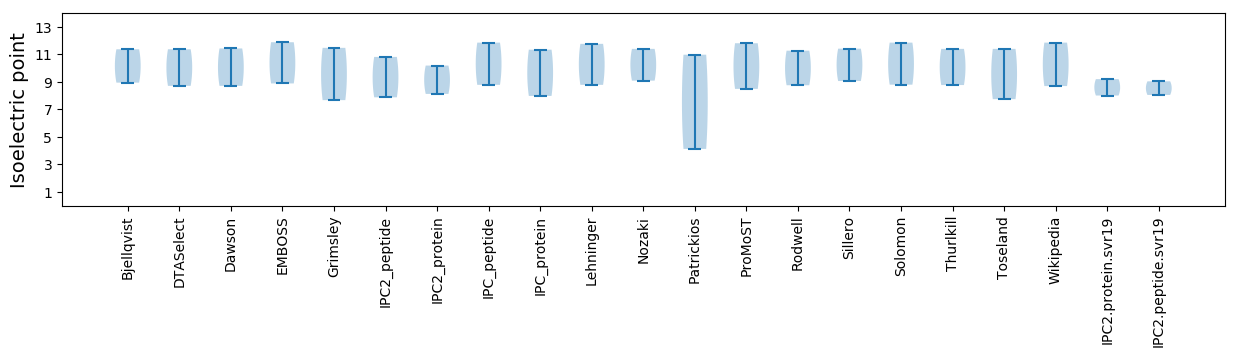
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D4N3R5|D4N3R5_9CIRC Capsid protein OS=Cyclovirus TN18 OX=742925 GN=cap PE=4 SV=1
MM1 pKa = 7.67AFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84VGRR9 pKa = 11.84SRR11 pKa = 11.84APIRR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 9.68LPRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84VARR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84FRR30 pKa = 11.84RR31 pKa = 11.84SRR33 pKa = 11.84RR34 pKa = 11.84LRR36 pKa = 11.84GNFTVLAKK44 pKa = 9.39RR45 pKa = 11.84TQVVVIQGDD54 pKa = 3.39EE55 pKa = 4.3GGTVTIAPSLNDD67 pKa = 3.49FTEE70 pKa = 4.49LAPLVPNFEE79 pKa = 4.5GFRR82 pKa = 11.84IWSVSCKK89 pKa = 9.6IRR91 pKa = 11.84PLFNVASDD99 pKa = 3.42VGPVPRR105 pKa = 11.84YY106 pKa = 9.65YY107 pKa = 10.33VAPWHH112 pKa = 6.69KK113 pKa = 8.47PTPTTVDD120 pKa = 3.37SNGVLSIDD128 pKa = 3.84RR129 pKa = 11.84SKK131 pKa = 11.25SYY133 pKa = 11.13NGTSGAFRR141 pKa = 11.84RR142 pKa = 11.84FVPALLSAVGYY153 pKa = 10.4SGIAGNQYY161 pKa = 10.94GKK163 pKa = 9.99IEE165 pKa = 3.73WRR167 pKa = 11.84PRR169 pKa = 11.84VEE171 pKa = 4.79LNSNTQSLQHH181 pKa = 5.61YY182 pKa = 8.88CGVVHH187 pKa = 6.96WSKK190 pKa = 11.26DD191 pKa = 3.51QLPGAGAPSRR201 pKa = 11.84RR202 pKa = 11.84QYY204 pKa = 10.68EE205 pKa = 3.74IEE207 pKa = 4.86LIAKK211 pKa = 7.61ITLYY215 pKa = 8.74NQKK218 pKa = 10.26YY219 pKa = 10.06FIGG222 pKa = 3.96
MM1 pKa = 7.67AFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84VGRR9 pKa = 11.84SRR11 pKa = 11.84APIRR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 9.68LPRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84VARR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84FRR30 pKa = 11.84RR31 pKa = 11.84SRR33 pKa = 11.84RR34 pKa = 11.84LRR36 pKa = 11.84GNFTVLAKK44 pKa = 9.39RR45 pKa = 11.84TQVVVIQGDD54 pKa = 3.39EE55 pKa = 4.3GGTVTIAPSLNDD67 pKa = 3.49FTEE70 pKa = 4.49LAPLVPNFEE79 pKa = 4.5GFRR82 pKa = 11.84IWSVSCKK89 pKa = 9.6IRR91 pKa = 11.84PLFNVASDD99 pKa = 3.42VGPVPRR105 pKa = 11.84YY106 pKa = 9.65YY107 pKa = 10.33VAPWHH112 pKa = 6.69KK113 pKa = 8.47PTPTTVDD120 pKa = 3.37SNGVLSIDD128 pKa = 3.84RR129 pKa = 11.84SKK131 pKa = 11.25SYY133 pKa = 11.13NGTSGAFRR141 pKa = 11.84RR142 pKa = 11.84FVPALLSAVGYY153 pKa = 10.4SGIAGNQYY161 pKa = 10.94GKK163 pKa = 9.99IEE165 pKa = 3.73WRR167 pKa = 11.84PRR169 pKa = 11.84VEE171 pKa = 4.79LNSNTQSLQHH181 pKa = 5.61YY182 pKa = 8.88CGVVHH187 pKa = 6.96WSKK190 pKa = 11.26DD191 pKa = 3.51QLPGAGAPSRR201 pKa = 11.84RR202 pKa = 11.84QYY204 pKa = 10.68EE205 pKa = 3.74IEE207 pKa = 4.86LIAKK211 pKa = 7.61ITLYY215 pKa = 8.74NQKK218 pKa = 10.26YY219 pKa = 10.06FIGG222 pKa = 3.96
Molecular weight: 25.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
508 |
222 |
286 |
254.0 |
29.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.709 ± 0.675 | 2.362 ± 0.941 |
3.543 ± 0.541 | 5.709 ± 1.646 |
4.724 ± 0.142 | 8.071 ± 0.024 |
1.575 ± 0.144 | 6.102 ± 0.449 |
6.102 ± 1.319 | 6.299 ± 0.295 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.591 ± 0.09 | 4.134 ± 0.239 |
4.921 ± 1.182 | 3.74 ± 0.088 |
10.039 ± 2.238 | 7.087 ± 0.368 |
5.709 ± 0.485 | 6.102 ± 1.872 |
2.165 ± 0.234 | 5.315 ± 0.812 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
