
Halioglobus lutimaris
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; Halioglobus
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
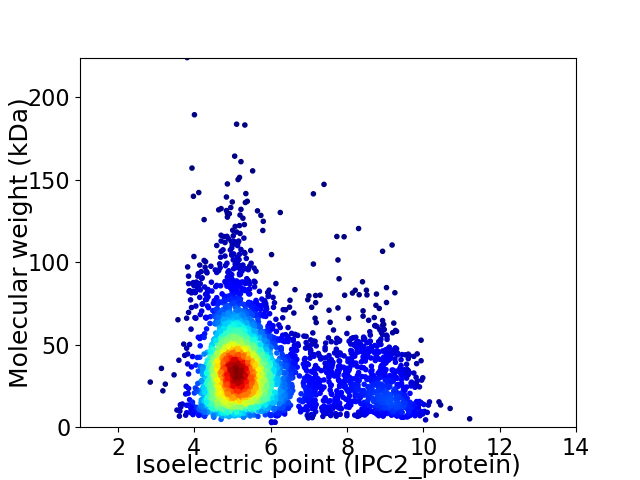
Virtual 2D-PAGE plot for 4080 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N5X847|A0A2N5X847_9GAMM N-acetyltransferase domain-containing protein OS=Halioglobus lutimaris OX=1737061 GN=C0039_00520 PE=4 SV=1
MM1 pKa = 7.29SVNLIVANDD10 pKa = 3.47DD11 pKa = 4.05SFFHH15 pKa = 6.65EE16 pKa = 4.23ARR18 pKa = 11.84SIKK21 pKa = 10.22EE22 pKa = 4.01NNTEE26 pKa = 3.71EE27 pKa = 6.08AIMFKK32 pKa = 10.84PEE34 pKa = 3.95FKK36 pKa = 10.21LTLLSAAISTFIPLNLADD54 pKa = 4.59AATGCEE60 pKa = 3.79NSARR64 pKa = 11.84LMMRR68 pKa = 11.84SCVAEE73 pKa = 3.93VRR75 pKa = 11.84EE76 pKa = 4.24EE77 pKa = 4.06LTATLAACQSEE88 pKa = 4.86SNNAIRR94 pKa = 11.84KK95 pKa = 7.49ACRR98 pKa = 11.84SAAQEE103 pKa = 4.04EE104 pKa = 4.65VAEE107 pKa = 4.98ARR109 pKa = 11.84DD110 pKa = 3.55TCQEE114 pKa = 3.32QRR116 pKa = 11.84AARR119 pKa = 11.84RR120 pKa = 11.84DD121 pKa = 3.41ACDD124 pKa = 3.32EE125 pKa = 4.25LNEE128 pKa = 4.89DD129 pKa = 4.65RR130 pKa = 11.84YY131 pKa = 11.52DD132 pKa = 3.97DD133 pKa = 4.37PLVDD137 pKa = 4.11GSIKK141 pKa = 10.48FVDD144 pKa = 4.17PDD146 pKa = 5.04DD147 pKa = 6.15IGDD150 pKa = 3.66TWPNNPYY157 pKa = 10.58VILQAGHH164 pKa = 4.9THH166 pKa = 5.77VLAAEE171 pKa = 4.16DD172 pKa = 4.51EE173 pKa = 4.56IVVVYY178 pKa = 8.83ATEE181 pKa = 4.61EE182 pKa = 3.52IRR184 pKa = 11.84EE185 pKa = 4.17IQGVPCRR192 pKa = 11.84VVADD196 pKa = 3.43VVVEE200 pKa = 3.88EE201 pKa = 5.04EE202 pKa = 4.16YY203 pKa = 11.2DD204 pKa = 3.84DD205 pKa = 6.1EE206 pKa = 4.77EE207 pKa = 5.41GEE209 pKa = 3.87WDD211 pKa = 3.56YY212 pKa = 11.8TPIEE216 pKa = 4.27VTDD219 pKa = 3.57DD220 pKa = 3.07WFAQDD225 pKa = 3.55EE226 pKa = 4.45NSNVYY231 pKa = 10.31YY232 pKa = 10.39CGEE235 pKa = 3.87VAQNFEE241 pKa = 4.65DD242 pKa = 4.64GVLRR246 pKa = 11.84DD247 pKa = 3.8LDD249 pKa = 3.88GSFEE253 pKa = 4.33SGLDD257 pKa = 3.36RR258 pKa = 11.84AKK260 pKa = 11.09GGLLTLAMPSPGDD273 pKa = 3.24IHH275 pKa = 6.91RR276 pKa = 11.84QEE278 pKa = 4.29YY279 pKa = 10.12FLGEE283 pKa = 4.16AEE285 pKa = 6.15DD286 pKa = 3.56IVQYY290 pKa = 11.19LDD292 pKa = 4.71LSASPSEE299 pKa = 4.26EE300 pKa = 3.42EE301 pKa = 4.09GGEE304 pKa = 4.07NEE306 pKa = 5.58AYY308 pKa = 9.27PCQGNCLKK316 pKa = 10.04TFDD319 pKa = 4.38FAPLEE324 pKa = 4.45PEE326 pKa = 3.78STEE329 pKa = 3.71YY330 pKa = 10.58KK331 pKa = 10.52YY332 pKa = 10.9YY333 pKa = 10.96LPGTGFVLAVSLEE346 pKa = 4.17DD347 pKa = 4.08GEE349 pKa = 4.22VDD351 pKa = 3.4EE352 pKa = 5.38EE353 pKa = 4.45GRR355 pKa = 11.84EE356 pKa = 4.18SLVCAGPSLEE366 pKa = 4.34VLFDD370 pKa = 4.08DD371 pKa = 4.79ACGIEE376 pKa = 4.55DD377 pKa = 4.41PEE379 pKa = 4.18EE380 pKa = 4.12LMEE383 pKa = 5.47DD384 pKa = 2.89LCEE387 pKa = 4.11FHH389 pKa = 7.74NEE391 pKa = 3.68FCEE394 pKa = 5.03DD395 pKa = 4.61DD396 pKa = 3.85EE397 pKa = 6.75DD398 pKa = 4.54GQQ400 pKa = 4.23
MM1 pKa = 7.29SVNLIVANDD10 pKa = 3.47DD11 pKa = 4.05SFFHH15 pKa = 6.65EE16 pKa = 4.23ARR18 pKa = 11.84SIKK21 pKa = 10.22EE22 pKa = 4.01NNTEE26 pKa = 3.71EE27 pKa = 6.08AIMFKK32 pKa = 10.84PEE34 pKa = 3.95FKK36 pKa = 10.21LTLLSAAISTFIPLNLADD54 pKa = 4.59AATGCEE60 pKa = 3.79NSARR64 pKa = 11.84LMMRR68 pKa = 11.84SCVAEE73 pKa = 3.93VRR75 pKa = 11.84EE76 pKa = 4.24EE77 pKa = 4.06LTATLAACQSEE88 pKa = 4.86SNNAIRR94 pKa = 11.84KK95 pKa = 7.49ACRR98 pKa = 11.84SAAQEE103 pKa = 4.04EE104 pKa = 4.65VAEE107 pKa = 4.98ARR109 pKa = 11.84DD110 pKa = 3.55TCQEE114 pKa = 3.32QRR116 pKa = 11.84AARR119 pKa = 11.84RR120 pKa = 11.84DD121 pKa = 3.41ACDD124 pKa = 3.32EE125 pKa = 4.25LNEE128 pKa = 4.89DD129 pKa = 4.65RR130 pKa = 11.84YY131 pKa = 11.52DD132 pKa = 3.97DD133 pKa = 4.37PLVDD137 pKa = 4.11GSIKK141 pKa = 10.48FVDD144 pKa = 4.17PDD146 pKa = 5.04DD147 pKa = 6.15IGDD150 pKa = 3.66TWPNNPYY157 pKa = 10.58VILQAGHH164 pKa = 4.9THH166 pKa = 5.77VLAAEE171 pKa = 4.16DD172 pKa = 4.51EE173 pKa = 4.56IVVVYY178 pKa = 8.83ATEE181 pKa = 4.61EE182 pKa = 3.52IRR184 pKa = 11.84EE185 pKa = 4.17IQGVPCRR192 pKa = 11.84VVADD196 pKa = 3.43VVVEE200 pKa = 3.88EE201 pKa = 5.04EE202 pKa = 4.16YY203 pKa = 11.2DD204 pKa = 3.84DD205 pKa = 6.1EE206 pKa = 4.77EE207 pKa = 5.41GEE209 pKa = 3.87WDD211 pKa = 3.56YY212 pKa = 11.8TPIEE216 pKa = 4.27VTDD219 pKa = 3.57DD220 pKa = 3.07WFAQDD225 pKa = 3.55EE226 pKa = 4.45NSNVYY231 pKa = 10.31YY232 pKa = 10.39CGEE235 pKa = 3.87VAQNFEE241 pKa = 4.65DD242 pKa = 4.64GVLRR246 pKa = 11.84DD247 pKa = 3.8LDD249 pKa = 3.88GSFEE253 pKa = 4.33SGLDD257 pKa = 3.36RR258 pKa = 11.84AKK260 pKa = 11.09GGLLTLAMPSPGDD273 pKa = 3.24IHH275 pKa = 6.91RR276 pKa = 11.84QEE278 pKa = 4.29YY279 pKa = 10.12FLGEE283 pKa = 4.16AEE285 pKa = 6.15DD286 pKa = 3.56IVQYY290 pKa = 11.19LDD292 pKa = 4.71LSASPSEE299 pKa = 4.26EE300 pKa = 3.42EE301 pKa = 4.09GGEE304 pKa = 4.07NEE306 pKa = 5.58AYY308 pKa = 9.27PCQGNCLKK316 pKa = 10.04TFDD319 pKa = 4.38FAPLEE324 pKa = 4.45PEE326 pKa = 3.78STEE329 pKa = 3.71YY330 pKa = 10.58KK331 pKa = 10.52YY332 pKa = 10.9YY333 pKa = 10.96LPGTGFVLAVSLEE346 pKa = 4.17DD347 pKa = 4.08GEE349 pKa = 4.22VDD351 pKa = 3.4EE352 pKa = 5.38EE353 pKa = 4.45GRR355 pKa = 11.84EE356 pKa = 4.18SLVCAGPSLEE366 pKa = 4.34VLFDD370 pKa = 4.08DD371 pKa = 4.79ACGIEE376 pKa = 4.55DD377 pKa = 4.41PEE379 pKa = 4.18EE380 pKa = 4.12LMEE383 pKa = 5.47DD384 pKa = 2.89LCEE387 pKa = 4.11FHH389 pKa = 7.74NEE391 pKa = 3.68FCEE394 pKa = 5.03DD395 pKa = 4.61DD396 pKa = 3.85EE397 pKa = 6.75DD398 pKa = 4.54GQQ400 pKa = 4.23
Molecular weight: 44.44 kDa
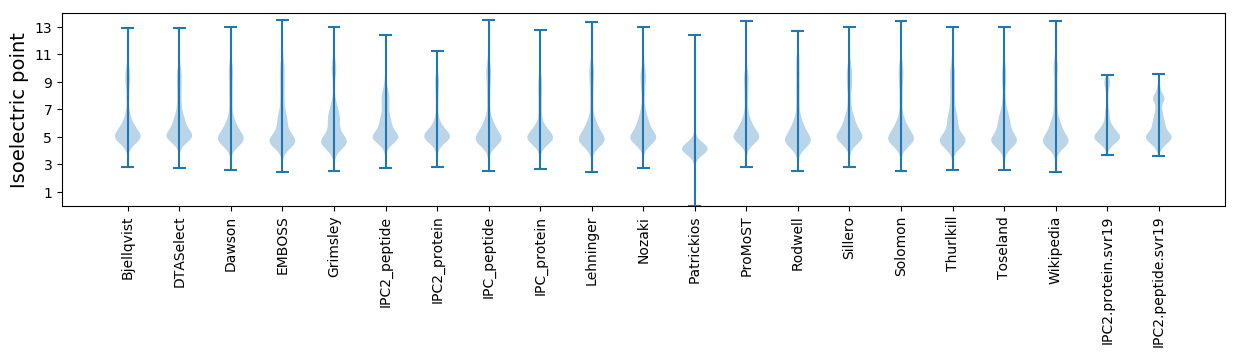
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N5X8V7|A0A2N5X8V7_9GAMM SPOR domain-containing protein OS=Halioglobus lutimaris OX=1737061 GN=C0039_02030 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.87RR14 pKa = 11.84THH16 pKa = 5.99GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.44GGRR28 pKa = 11.84AVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84GKK37 pKa = 9.75GRR39 pKa = 11.84KK40 pKa = 8.78VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.87RR14 pKa = 11.84THH16 pKa = 5.99GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.44GGRR28 pKa = 11.84AVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84GKK37 pKa = 9.75GRR39 pKa = 11.84KK40 pKa = 8.78VLSAA44 pKa = 4.05
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1394984 |
25 |
2139 |
341.9 |
37.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.493 ± 0.041 | 1.069 ± 0.014 |
5.905 ± 0.034 | 6.438 ± 0.035 |
3.671 ± 0.023 | 8.007 ± 0.034 |
2.182 ± 0.017 | 5.027 ± 0.028 |
3.262 ± 0.03 | 10.548 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.545 ± 0.017 | 3.176 ± 0.024 |
4.673 ± 0.023 | 4.082 ± 0.023 |
6.338 ± 0.031 | 6.101 ± 0.028 |
5.033 ± 0.021 | 7.229 ± 0.033 |
1.398 ± 0.017 | 2.822 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
