
Cylindrobasidium torrendii FP15055 ss-10
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Physalacriaceae; Cylindrobasidium; Cylindrobasidium torrendii
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
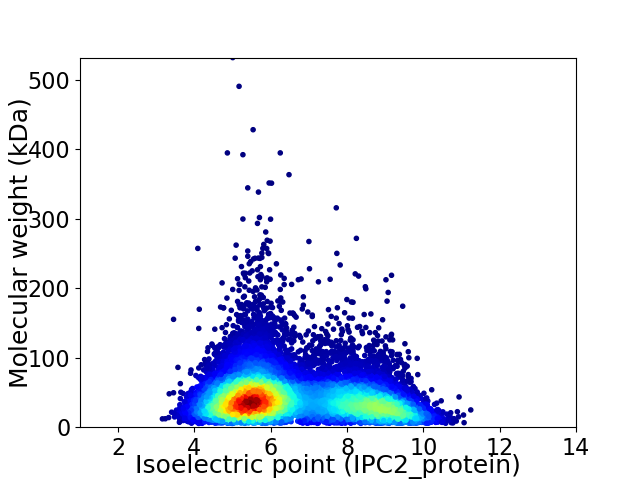
Virtual 2D-PAGE plot for 13901 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D7BB58|A0A0D7BB58_9AGAR Uncharacterized protein OS=Cylindrobasidium torrendii FP15055 ss-10 OX=1314674 GN=CYLTODRAFT_454512 PE=4 SV=1
MM1 pKa = 7.17FALAALSLTVLPFVSADD18 pKa = 3.48TALDD22 pKa = 3.6LAGIKK27 pKa = 10.15AHH29 pKa = 6.02FTQSKK34 pKa = 9.54IVPDD38 pKa = 4.1LFEE41 pKa = 4.59SFNPEE46 pKa = 3.03AVLTVNFPGVGDD58 pKa = 3.72ITPGQTLTQAEE69 pKa = 4.31AGGYY73 pKa = 9.54PSIKK77 pKa = 8.77VTAANSSVDD86 pKa = 3.58LGSTFTVAMVDD97 pKa = 3.33ASYY100 pKa = 11.23VGQDD104 pKa = 3.43DD105 pKa = 5.55DD106 pKa = 5.19DD107 pKa = 4.31VTRR110 pKa = 11.84HH111 pKa = 5.15WLMNGCTLSDD121 pKa = 3.48GTISNSSATSITEE134 pKa = 3.96YY135 pKa = 10.72GGPAPPEE142 pKa = 4.11GEE144 pKa = 3.76PAHH147 pKa = 6.66RR148 pKa = 11.84YY149 pKa = 9.2VIAVYY154 pKa = 8.16QQPDD158 pKa = 3.63SFSAPEE164 pKa = 4.44GFNQANMGISTFDD177 pKa = 3.47FEE179 pKa = 6.83DD180 pKa = 3.91YY181 pKa = 9.67LTNSKK186 pKa = 10.54LGALVGATYY195 pKa = 7.7MTVQTGEE202 pKa = 3.86ATTSVEE208 pKa = 4.1ATSSVDD214 pKa = 3.49SQTLSAASQTGSSSSGASATTGASGSSSSGAPDD247 pKa = 3.49GASALTASSAAVALGFLGFLFAA269 pKa = 6.43
MM1 pKa = 7.17FALAALSLTVLPFVSADD18 pKa = 3.48TALDD22 pKa = 3.6LAGIKK27 pKa = 10.15AHH29 pKa = 6.02FTQSKK34 pKa = 9.54IVPDD38 pKa = 4.1LFEE41 pKa = 4.59SFNPEE46 pKa = 3.03AVLTVNFPGVGDD58 pKa = 3.72ITPGQTLTQAEE69 pKa = 4.31AGGYY73 pKa = 9.54PSIKK77 pKa = 8.77VTAANSSVDD86 pKa = 3.58LGSTFTVAMVDD97 pKa = 3.33ASYY100 pKa = 11.23VGQDD104 pKa = 3.43DD105 pKa = 5.55DD106 pKa = 5.19DD107 pKa = 4.31VTRR110 pKa = 11.84HH111 pKa = 5.15WLMNGCTLSDD121 pKa = 3.48GTISNSSATSITEE134 pKa = 3.96YY135 pKa = 10.72GGPAPPEE142 pKa = 4.11GEE144 pKa = 3.76PAHH147 pKa = 6.66RR148 pKa = 11.84YY149 pKa = 9.2VIAVYY154 pKa = 8.16QQPDD158 pKa = 3.63SFSAPEE164 pKa = 4.44GFNQANMGISTFDD177 pKa = 3.47FEE179 pKa = 6.83DD180 pKa = 3.91YY181 pKa = 9.67LTNSKK186 pKa = 10.54LGALVGATYY195 pKa = 7.7MTVQTGEE202 pKa = 3.86ATTSVEE208 pKa = 4.1ATSSVDD214 pKa = 3.49SQTLSAASQTGSSSSGASATTGASGSSSSGAPDD247 pKa = 3.49GASALTASSAAVALGFLGFLFAA269 pKa = 6.43
Molecular weight: 27.21 kDa
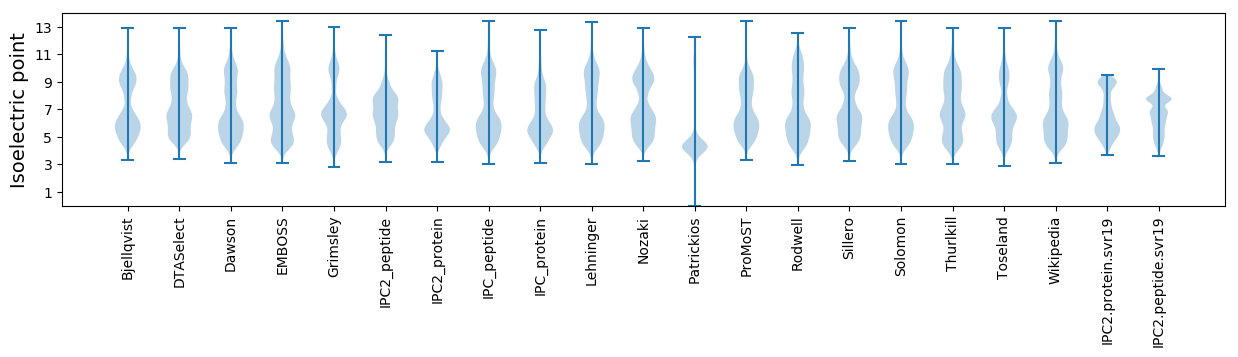
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D7B3P3|A0A0D7B3P3_9AGAR Ribosomal protein OS=Cylindrobasidium torrendii FP15055 ss-10 OX=1314674 GN=CYLTODRAFT_456859 PE=3 SV=1
CC1 pKa = 7.47LLAKK5 pKa = 10.53LLLRR9 pKa = 11.84TPTSPRR15 pKa = 11.84VVLRR19 pKa = 11.84ASRR22 pKa = 11.84SSRR25 pKa = 11.84SQLKK29 pKa = 10.05SPRR32 pKa = 11.84RR33 pKa = 11.84PPRR36 pKa = 11.84NPAPKK41 pKa = 10.16KK42 pKa = 9.71PIRR45 pKa = 11.84LRR47 pKa = 11.84TARR50 pKa = 11.84RR51 pKa = 11.84KK52 pKa = 9.25PRR54 pKa = 11.84NLRR57 pKa = 11.84ARR59 pKa = 11.84SARR62 pKa = 11.84LLKK65 pKa = 9.92TLTKK69 pKa = 9.7PPSPRR74 pKa = 11.84RR75 pKa = 11.84PNPPAKK81 pKa = 9.98RR82 pKa = 11.84SLLPRR87 pKa = 11.84QSPPARR93 pKa = 11.84LSQRR97 pKa = 11.84PRR99 pKa = 11.84QHH101 pKa = 4.75QHH103 pKa = 6.22RR104 pKa = 11.84SLHH107 pKa = 5.82PALLRR112 pKa = 11.84PSQRR116 pKa = 11.84LVLLLSQRR124 pKa = 11.84LGLRR128 pKa = 11.84QQSLPLVPPPRR139 pKa = 11.84NLHH142 pKa = 5.96RR143 pKa = 11.84ALGPRR148 pKa = 11.84SRR150 pKa = 11.84HH151 pKa = 5.15PRR153 pKa = 11.84LVRR156 pKa = 11.84PPLPLLRR163 pKa = 11.84PPPRR167 pKa = 11.84RR168 pKa = 11.84QSLKK172 pKa = 10.59SRR174 pKa = 11.84NPLKK178 pKa = 9.71KK179 pKa = 8.88TKK181 pKa = 9.98LRR183 pKa = 11.84PPLKK187 pKa = 8.14TRR189 pKa = 11.84PRR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84CPRR196 pKa = 11.84RR197 pKa = 11.84RR198 pKa = 11.84KK199 pKa = 9.56PEE201 pKa = 3.76NGRR204 pKa = 11.84GWWAIGYY211 pKa = 9.39RR212 pKa = 11.84LL213 pKa = 3.88
CC1 pKa = 7.47LLAKK5 pKa = 10.53LLLRR9 pKa = 11.84TPTSPRR15 pKa = 11.84VVLRR19 pKa = 11.84ASRR22 pKa = 11.84SSRR25 pKa = 11.84SQLKK29 pKa = 10.05SPRR32 pKa = 11.84RR33 pKa = 11.84PPRR36 pKa = 11.84NPAPKK41 pKa = 10.16KK42 pKa = 9.71PIRR45 pKa = 11.84LRR47 pKa = 11.84TARR50 pKa = 11.84RR51 pKa = 11.84KK52 pKa = 9.25PRR54 pKa = 11.84NLRR57 pKa = 11.84ARR59 pKa = 11.84SARR62 pKa = 11.84LLKK65 pKa = 9.92TLTKK69 pKa = 9.7PPSPRR74 pKa = 11.84RR75 pKa = 11.84PNPPAKK81 pKa = 9.98RR82 pKa = 11.84SLLPRR87 pKa = 11.84QSPPARR93 pKa = 11.84LSQRR97 pKa = 11.84PRR99 pKa = 11.84QHH101 pKa = 4.75QHH103 pKa = 6.22RR104 pKa = 11.84SLHH107 pKa = 5.82PALLRR112 pKa = 11.84PSQRR116 pKa = 11.84LVLLLSQRR124 pKa = 11.84LGLRR128 pKa = 11.84QQSLPLVPPPRR139 pKa = 11.84NLHH142 pKa = 5.96RR143 pKa = 11.84ALGPRR148 pKa = 11.84SRR150 pKa = 11.84HH151 pKa = 5.15PRR153 pKa = 11.84LVRR156 pKa = 11.84PPLPLLRR163 pKa = 11.84PPPRR167 pKa = 11.84RR168 pKa = 11.84QSLKK172 pKa = 10.59SRR174 pKa = 11.84NPLKK178 pKa = 9.71KK179 pKa = 8.88TKK181 pKa = 9.98LRR183 pKa = 11.84PPLKK187 pKa = 8.14TRR189 pKa = 11.84PRR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84CPRR196 pKa = 11.84RR197 pKa = 11.84RR198 pKa = 11.84KK199 pKa = 9.56PEE201 pKa = 3.76NGRR204 pKa = 11.84GWWAIGYY211 pKa = 9.39RR212 pKa = 11.84LL213 pKa = 3.88
Molecular weight: 24.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5747761 |
50 |
4803 |
413.5 |
45.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.91 ± 0.019 | 1.314 ± 0.009 |
5.796 ± 0.016 | 5.907 ± 0.02 |
3.737 ± 0.013 | 6.414 ± 0.021 |
2.523 ± 0.008 | 4.729 ± 0.014 |
4.558 ± 0.019 | 9.116 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.234 ± 0.008 | 3.378 ± 0.011 |
6.436 ± 0.03 | 3.644 ± 0.013 |
6.276 ± 0.018 | 8.307 ± 0.026 |
6.073 ± 0.012 | 6.398 ± 0.014 |
1.509 ± 0.009 | 2.741 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
