
Psychrosphaera saromensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Pseudoalteromonadaceae; Psychrosphaera
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
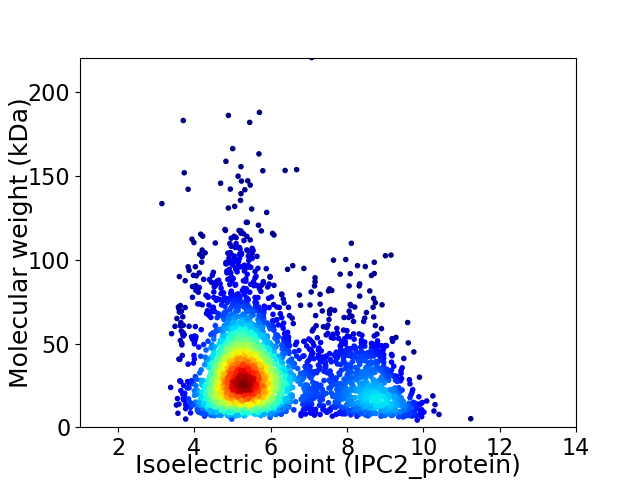
Virtual 2D-PAGE plot for 3139 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S7UZP6|A0A2S7UZP6_9GAMM dTDP-4-dehydrorhamnose reductase OS=Psychrosphaera saromensis OX=716813 GN=BTO11_14565 PE=3 SV=1
MM1 pKa = 7.93LSRR4 pKa = 11.84LRR6 pKa = 11.84NRR8 pKa = 11.84LIPINSALSTNLNSSSNVLSFSKK31 pKa = 10.64INFQMLDD38 pKa = 3.83LIKK41 pKa = 10.46RR42 pKa = 11.84SALIISTVFILAGCSSDD59 pKa = 4.12DD60 pKa = 3.59RR61 pKa = 11.84SSEE64 pKa = 4.02DD65 pKa = 3.18VVEE68 pKa = 4.22EE69 pKa = 4.39VEE71 pKa = 4.15AGNAVGYY78 pKa = 10.31VITVNGALIVDD89 pKa = 4.43WPQGFDD95 pKa = 3.83YY96 pKa = 10.67IDD98 pKa = 3.97AGAMVYY104 pKa = 10.71DD105 pKa = 4.2EE106 pKa = 5.99GGDD109 pKa = 3.51FYY111 pKa = 11.8VFTDD115 pKa = 3.56GDD117 pKa = 3.8SRR119 pKa = 11.84NIDD122 pKa = 3.26TSILGEE128 pKa = 4.08HH129 pKa = 5.99TVTFSLIDD137 pKa = 3.46SEE139 pKa = 4.97GNEE142 pKa = 4.22TVATRR147 pKa = 11.84TVNVFAPIPFITTWTTTEE165 pKa = 4.07SNQSIRR171 pKa = 11.84IPIYY175 pKa = 10.74SFNTYY180 pKa = 10.62YY181 pKa = 10.91DD182 pKa = 4.47FSIDD186 pKa = 3.47WGDD189 pKa = 3.44GSQIDD194 pKa = 4.07EE195 pKa = 4.39NQTDD199 pKa = 4.49SVTHH203 pKa = 6.74IYY205 pKa = 8.64STAGTYY211 pKa = 7.64TVSITGIFPHH221 pKa = 6.94MYY223 pKa = 9.66NDD225 pKa = 3.84EE226 pKa = 4.18NSSVDD231 pKa = 2.97KK232 pKa = 11.19LMSVEE237 pKa = 3.56QWGNIRR243 pKa = 11.84WSSMRR248 pKa = 11.84YY249 pKa = 6.27MFRR252 pKa = 11.84GAHH255 pKa = 5.54NLVINATDD263 pKa = 3.71APVLSDD269 pKa = 3.94VEE271 pKa = 4.35SMYY274 pKa = 11.69AMFYY278 pKa = 10.29GASSFNTDD286 pKa = 2.17ISHH289 pKa = 6.88WDD291 pKa = 3.44VSNVTDD297 pKa = 3.95MSVMFYY303 pKa = 10.84GAAAFNQPIDD313 pKa = 3.36GWDD316 pKa = 3.45VSNVTNMVWMFFNAAVFNQPLNSWDD341 pKa = 3.57VSNVTDD347 pKa = 3.77MSGMFYY353 pKa = 10.67VAAEE357 pKa = 4.22FNQPIDD363 pKa = 3.05SWDD366 pKa = 3.5ASNVTNMSFMFQLAEE381 pKa = 4.38SFNQPIDD388 pKa = 3.19SWDD391 pKa = 3.52VSNVTDD397 pKa = 3.89MSWMFGGTAAFNQPLDD413 pKa = 3.46SWDD416 pKa = 3.56VSNVTDD422 pKa = 3.71MSGMFAGAGVFNQPLDD438 pKa = 3.41SWDD441 pKa = 3.41VSRR444 pKa = 11.84VTNMSIMFHH453 pKa = 6.61RR454 pKa = 11.84LAAFNQPLDD463 pKa = 3.46SWDD466 pKa = 3.56VSNVTDD472 pKa = 3.96MEE474 pKa = 5.25SMFSGTTAFNQPLDD488 pKa = 3.54SWDD491 pKa = 3.58VSNVTTMINMLNSSGLSTDD510 pKa = 4.04NYY512 pKa = 11.16DD513 pKa = 3.61ALLNGWSIQILQPDD527 pKa = 4.91LIFDD531 pKa = 3.86VSATYY536 pKa = 10.57SSSSQAARR544 pKa = 11.84QSIIDD549 pKa = 3.82TYY551 pKa = 11.6NWTINDD557 pKa = 4.97DD558 pKa = 3.91GLQPP562 pKa = 4.09
MM1 pKa = 7.93LSRR4 pKa = 11.84LRR6 pKa = 11.84NRR8 pKa = 11.84LIPINSALSTNLNSSSNVLSFSKK31 pKa = 10.64INFQMLDD38 pKa = 3.83LIKK41 pKa = 10.46RR42 pKa = 11.84SALIISTVFILAGCSSDD59 pKa = 4.12DD60 pKa = 3.59RR61 pKa = 11.84SSEE64 pKa = 4.02DD65 pKa = 3.18VVEE68 pKa = 4.22EE69 pKa = 4.39VEE71 pKa = 4.15AGNAVGYY78 pKa = 10.31VITVNGALIVDD89 pKa = 4.43WPQGFDD95 pKa = 3.83YY96 pKa = 10.67IDD98 pKa = 3.97AGAMVYY104 pKa = 10.71DD105 pKa = 4.2EE106 pKa = 5.99GGDD109 pKa = 3.51FYY111 pKa = 11.8VFTDD115 pKa = 3.56GDD117 pKa = 3.8SRR119 pKa = 11.84NIDD122 pKa = 3.26TSILGEE128 pKa = 4.08HH129 pKa = 5.99TVTFSLIDD137 pKa = 3.46SEE139 pKa = 4.97GNEE142 pKa = 4.22TVATRR147 pKa = 11.84TVNVFAPIPFITTWTTTEE165 pKa = 4.07SNQSIRR171 pKa = 11.84IPIYY175 pKa = 10.74SFNTYY180 pKa = 10.62YY181 pKa = 10.91DD182 pKa = 4.47FSIDD186 pKa = 3.47WGDD189 pKa = 3.44GSQIDD194 pKa = 4.07EE195 pKa = 4.39NQTDD199 pKa = 4.49SVTHH203 pKa = 6.74IYY205 pKa = 8.64STAGTYY211 pKa = 7.64TVSITGIFPHH221 pKa = 6.94MYY223 pKa = 9.66NDD225 pKa = 3.84EE226 pKa = 4.18NSSVDD231 pKa = 2.97KK232 pKa = 11.19LMSVEE237 pKa = 3.56QWGNIRR243 pKa = 11.84WSSMRR248 pKa = 11.84YY249 pKa = 6.27MFRR252 pKa = 11.84GAHH255 pKa = 5.54NLVINATDD263 pKa = 3.71APVLSDD269 pKa = 3.94VEE271 pKa = 4.35SMYY274 pKa = 11.69AMFYY278 pKa = 10.29GASSFNTDD286 pKa = 2.17ISHH289 pKa = 6.88WDD291 pKa = 3.44VSNVTDD297 pKa = 3.95MSVMFYY303 pKa = 10.84GAAAFNQPIDD313 pKa = 3.36GWDD316 pKa = 3.45VSNVTNMVWMFFNAAVFNQPLNSWDD341 pKa = 3.57VSNVTDD347 pKa = 3.77MSGMFYY353 pKa = 10.67VAAEE357 pKa = 4.22FNQPIDD363 pKa = 3.05SWDD366 pKa = 3.5ASNVTNMSFMFQLAEE381 pKa = 4.38SFNQPIDD388 pKa = 3.19SWDD391 pKa = 3.52VSNVTDD397 pKa = 3.89MSWMFGGTAAFNQPLDD413 pKa = 3.46SWDD416 pKa = 3.56VSNVTDD422 pKa = 3.71MSGMFAGAGVFNQPLDD438 pKa = 3.41SWDD441 pKa = 3.41VSRR444 pKa = 11.84VTNMSIMFHH453 pKa = 6.61RR454 pKa = 11.84LAAFNQPLDD463 pKa = 3.46SWDD466 pKa = 3.56VSNVTDD472 pKa = 3.96MEE474 pKa = 5.25SMFSGTTAFNQPLDD488 pKa = 3.54SWDD491 pKa = 3.58VSNVTTMINMLNSSGLSTDD510 pKa = 4.04NYY512 pKa = 11.16DD513 pKa = 3.61ALLNGWSIQILQPDD527 pKa = 4.91LIFDD531 pKa = 3.86VSATYY536 pKa = 10.57SSSSQAARR544 pKa = 11.84QSIIDD549 pKa = 3.82TYY551 pKa = 11.6NWTINDD557 pKa = 4.97DD558 pKa = 3.91GLQPP562 pKa = 4.09
Molecular weight: 62.5 kDa
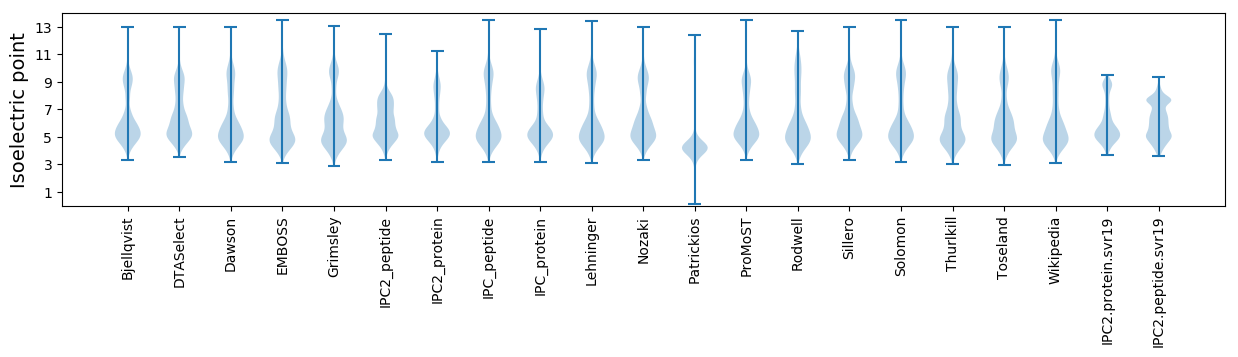
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S7USA2|A0A2S7USA2_9GAMM Dipeptidase OS=Psychrosphaera saromensis OX=716813 GN=BTO11_03480 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.39GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.39GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1031951 |
37 |
1982 |
328.8 |
36.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.835 ± 0.038 | 0.893 ± 0.014 |
5.885 ± 0.042 | 6.204 ± 0.042 |
4.399 ± 0.034 | 6.317 ± 0.04 |
1.99 ± 0.02 | 6.992 ± 0.036 |
6.231 ± 0.036 | 10.197 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.433 ± 0.021 | 5.112 ± 0.036 |
3.502 ± 0.027 | 4.397 ± 0.034 |
3.666 ± 0.028 | 7.08 ± 0.044 |
5.705 ± 0.031 | 6.866 ± 0.032 |
1.116 ± 0.018 | 3.18 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
