
Azospirillum sp. L-25-5w-1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Azospirillaceae; Azospirillum
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
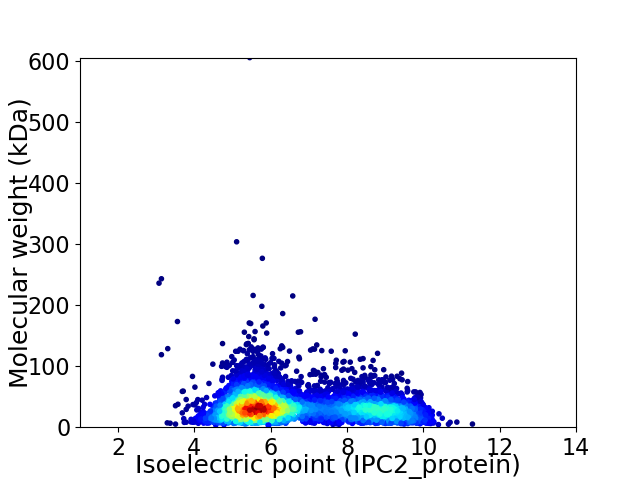
Virtual 2D-PAGE plot for 5160 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S0R6U4|A0A3S0R6U4_9PROT Dabb family protein OS=Azospirillum sp. L-25-5w-1 OX=2496639 GN=EJ903_19160 PE=4 SV=1
MM1 pKa = 7.62ASGISFLAGSTVKK14 pKa = 10.57SVNTYY19 pKa = 8.5TSSNQSYY26 pKa = 8.74PAIGSFNDD34 pKa = 3.38GGYY37 pKa = 8.4VVSWISYY44 pKa = 8.63LQDD47 pKa = 3.02GSGWGVYY54 pKa = 7.15GQRR57 pKa = 11.84FSTDD61 pKa = 4.16GIPVGGEE68 pKa = 3.26FRR70 pKa = 11.84INVSTTGDD78 pKa = 3.32QDD80 pKa = 3.63SPSIVTHH87 pKa = 6.48SDD89 pKa = 2.8GSFNVYY95 pKa = 7.51WASYY99 pKa = 7.98ATAGVPYY106 pKa = 8.13TTAANIYY113 pKa = 9.37GRR115 pKa = 11.84RR116 pKa = 11.84FDD118 pKa = 4.99ASGNPLGSEE127 pKa = 4.17FKK129 pKa = 10.98VNSATTSDD137 pKa = 3.56RR138 pKa = 11.84NRR140 pKa = 11.84VGTAKK145 pKa = 10.54LADD148 pKa = 3.97GGEE151 pKa = 4.39VVVWTLRR158 pKa = 11.84NAAWTTAEE166 pKa = 4.94LYY168 pKa = 10.5AQRR171 pKa = 11.84FDD173 pKa = 3.65SAGNSVGGEE182 pKa = 3.65FRR184 pKa = 11.84VNPTVTAASSGQPNSVAGLDD204 pKa = 3.56GGGFAVVWDD213 pKa = 4.22ASDD216 pKa = 3.97GNGSGVYY223 pKa = 7.74VQRR226 pKa = 11.84FDD228 pKa = 4.09ASGNRR233 pKa = 11.84VGMEE237 pKa = 3.66TRR239 pKa = 11.84VNSTVASDD247 pKa = 3.4QSMGNITALAGGGFVVSWTSNGQDD271 pKa = 3.07GGGTGVYY278 pKa = 9.81AQRR281 pKa = 11.84FDD283 pKa = 3.83AAGTPVGTEE292 pKa = 3.93FQVNTQTAANQYY304 pKa = 8.91GSSAIGSPDD313 pKa = 3.03GGFTILYY320 pKa = 6.96TAGDD324 pKa = 3.89NQDD327 pKa = 2.92GSGAGVYY334 pKa = 7.88GQRR337 pKa = 11.84YY338 pKa = 8.88DD339 pKa = 3.73SNGLRR344 pKa = 11.84VGGEE348 pKa = 3.63FRR350 pKa = 11.84VNDD353 pKa = 3.86STVGNQDD360 pKa = 2.86AQVIGVRR367 pKa = 11.84PDD369 pKa = 2.93GGFVVAWNSPDD380 pKa = 3.43ASQSGVLTRR389 pKa = 11.84IYY391 pKa = 10.88SNPTAPYY398 pKa = 9.89SVAGEE403 pKa = 3.9QRR405 pKa = 11.84VNTTGTGDD413 pKa = 3.4QTRR416 pKa = 11.84SSISVFADD424 pKa = 3.34GGYY427 pKa = 9.8VVTWVSAGQAGSGDD441 pKa = 3.3GVYY444 pKa = 10.46AQRR447 pKa = 11.84YY448 pKa = 8.61DD449 pKa = 3.65SGGKK453 pKa = 9.42PIGSEE458 pKa = 3.86FRR460 pKa = 11.84VNTTTTGDD468 pKa = 3.29QGDD471 pKa = 4.3ATVVAYY477 pKa = 10.37DD478 pKa = 4.03GGQFAIFWASHH489 pKa = 4.15TQAGQTSPVDD499 pKa = 3.25ASVRR503 pKa = 11.84GQRR506 pKa = 11.84YY507 pKa = 8.4AADD510 pKa = 3.77GTPVGSEE517 pKa = 3.94FQVNGASTSDD527 pKa = 3.62RR528 pKa = 11.84NPPGAVRR535 pKa = 11.84LADD538 pKa = 3.67GSTVVVWLQHH548 pKa = 5.94DD549 pKa = 4.31SSWAHH554 pKa = 4.28TWVYY558 pKa = 8.8GQRR561 pKa = 11.84YY562 pKa = 8.46DD563 pKa = 3.43AAGNAVGGEE572 pKa = 3.93FLVNSVNSANSGYY585 pKa = 9.95VASGYY590 pKa = 10.29QPASVTALEE599 pKa = 4.3NGGFAVVWNAADD611 pKa = 3.86GDD613 pKa = 4.14GTGAYY618 pKa = 8.22FQRR621 pKa = 11.84FDD623 pKa = 3.33ASANRR628 pKa = 11.84VGGEE632 pKa = 3.75VRR634 pKa = 11.84LNTTTASDD642 pKa = 3.32QFTASIAGLSGGGFVATWTSIGQDD666 pKa = 2.65GGTYY670 pKa = 10.33GIYY673 pKa = 10.38AQRR676 pKa = 11.84FDD678 pKa = 3.64SAGNAVGGEE687 pKa = 3.94FRR689 pKa = 11.84VNTITQGDD697 pKa = 3.98QYY699 pKa = 11.11ASRR702 pKa = 11.84VASLPDD708 pKa = 3.24GGFVVAYY715 pKa = 9.32SSSGGEE721 pKa = 4.11DD722 pKa = 3.3GSGIGVFGQRR732 pKa = 11.84YY733 pKa = 8.12DD734 pKa = 3.61AQGTAVGSEE743 pKa = 4.38FIITDD748 pKa = 3.58TTVGNQEE755 pKa = 4.13EE756 pKa = 4.38PHH758 pKa = 5.88LAARR762 pKa = 11.84PDD764 pKa = 3.48GGFVLTWVSPDD775 pKa = 3.0GNGRR779 pKa = 11.84GVFSRR784 pKa = 11.84LVRR787 pKa = 11.84AMAPPSATQAGTAGNDD803 pKa = 3.46LLTPANPRR811 pKa = 11.84SVLLGLAGNDD821 pKa = 3.48TLTGGSLTDD830 pKa = 3.66TLNGGDD836 pKa = 4.46GNDD839 pKa = 3.82SLSGGLGDD847 pKa = 5.22DD848 pKa = 3.92VLFGGIGNDD857 pKa = 3.5TLDD860 pKa = 3.87GGGGADD866 pKa = 4.36LLYY869 pKa = 11.08GGAGNDD875 pKa = 3.47TFIVRR880 pKa = 11.84GGEE883 pKa = 3.99ALSDD887 pKa = 3.21TGGIDD892 pKa = 3.37TAILTGPASYY902 pKa = 9.26TLPGGIEE909 pKa = 4.02NLVLTSAATGTGNSLDD925 pKa = 3.75NLLTGSSGSDD935 pKa = 3.05QLFGLAGNDD944 pKa = 3.56TLDD947 pKa = 3.8GGGGADD953 pKa = 3.67TLIGGLGNDD962 pKa = 3.89TYY964 pKa = 11.11IVDD967 pKa = 4.98DD968 pKa = 4.13IGDD971 pKa = 3.87VVTEE975 pKa = 4.23LSGEE979 pKa = 4.18GADD982 pKa = 4.04EE983 pKa = 4.19VRR985 pKa = 11.84TTLASYY991 pKa = 9.4TLGTNLEE998 pKa = 4.23NLTFTGSGSFTGTGNGLANILQGGAGNDD1026 pKa = 3.81TLDD1029 pKa = 3.78GGAGIDD1035 pKa = 3.6TLRR1038 pKa = 11.84GGVGNDD1044 pKa = 3.19TYY1046 pKa = 11.34LVSDD1050 pKa = 3.76VGDD1053 pKa = 3.84VIIEE1057 pKa = 4.1AVGEE1061 pKa = 4.3GTDD1064 pKa = 3.72EE1065 pKa = 4.28VRR1067 pKa = 11.84TTLASYY1073 pKa = 9.03TLAANVEE1080 pKa = 4.23NLVYY1084 pKa = 10.02TGTVAFTGTGNTGNNAITGGAGNDD1108 pKa = 3.95SLTGDD1113 pKa = 4.3AGNDD1117 pKa = 3.69TLDD1120 pKa = 3.9GGAGADD1126 pKa = 3.6TLVGGTGNDD1135 pKa = 3.43TYY1137 pKa = 11.11IVDD1140 pKa = 3.72TAGDD1144 pKa = 4.07VVTEE1148 pKa = 4.23LAGQGTDD1155 pKa = 3.15TVRR1158 pKa = 11.84TSLASYY1164 pKa = 9.13TLGANVEE1171 pKa = 4.28NLVFTGSGPVTFTGNALNNNLTGGAGADD1199 pKa = 4.07LLTGLDD1205 pKa = 4.67GNDD1208 pKa = 3.75TLNGGAGADD1217 pKa = 3.71TLLGGTGNDD1226 pKa = 3.28TYY1228 pKa = 11.76VVDD1231 pKa = 3.75NVGDD1235 pKa = 3.89VLVEE1239 pKa = 3.98LPGEE1243 pKa = 4.65GIDD1246 pKa = 3.89TVQSSINHH1254 pKa = 5.77TLLGDD1259 pKa = 3.98FEE1261 pKa = 5.41NLTLTGSAALTGTGNALNNLLTGNGAANLLTGLEE1295 pKa = 4.45GDD1297 pKa = 3.88DD1298 pKa = 3.88TLNGGGGADD1307 pKa = 3.62TLIGGLGNDD1316 pKa = 3.84SYY1318 pKa = 12.11VVDD1321 pKa = 3.72NAGDD1325 pKa = 3.74VVTEE1329 pKa = 3.96LVGEE1333 pKa = 4.83GIDD1336 pKa = 3.63TVNASINWTLGTNLEE1351 pKa = 4.13NLTLTGSAAISATGNGLNNLLTGNSGANLLDD1382 pKa = 4.28GGLGADD1388 pKa = 4.05SMVGGAGNDD1397 pKa = 3.43VYY1399 pKa = 11.0IVDD1402 pKa = 4.56DD1403 pKa = 4.64ADD1405 pKa = 4.04DD1406 pKa = 3.92VVTEE1410 pKa = 4.42LVGQGTDD1417 pKa = 3.14EE1418 pKa = 3.99VRR1420 pKa = 11.84TNLSSYY1426 pKa = 11.33LLGTNIEE1433 pKa = 4.44ALTHH1437 pKa = 6.16TGTGDD1442 pKa = 3.36FAGTGNALDD1451 pKa = 3.89NLLTGGLGNDD1461 pKa = 3.86TLSGGDD1467 pKa = 3.84GADD1470 pKa = 3.71TLDD1473 pKa = 4.34GGAGSNLLIGGTGHH1487 pKa = 7.61DD1488 pKa = 3.83VYY1490 pKa = 11.35LVGGFGDD1497 pKa = 3.54QVVEE1501 pKa = 3.93LVGEE1505 pKa = 4.35GMDD1508 pKa = 3.63EE1509 pKa = 4.06VRR1511 pKa = 11.84TALSGYY1517 pKa = 7.4TLTDD1521 pKa = 3.37GVEE1524 pKa = 4.1TLTYY1528 pKa = 9.09TGVAAFVGTGNGLDD1542 pKa = 3.56NLIAGGAGADD1552 pKa = 3.82TLDD1555 pKa = 4.19GGLGADD1561 pKa = 3.88TLIGGLGDD1569 pKa = 3.6DD1570 pKa = 4.24VYY1572 pKa = 11.74VVDD1575 pKa = 4.37SAGDD1579 pKa = 3.77TVIEE1583 pKa = 4.42GADD1586 pKa = 3.23AGLDD1590 pKa = 3.6EE1591 pKa = 5.1VRR1593 pKa = 11.84TTLSAFALGDD1603 pKa = 3.34NTEE1606 pKa = 4.05RR1607 pKa = 11.84LVFQGSGDD1615 pKa = 3.32ASGIGNGLDD1624 pKa = 2.81NWLAGNIGNDD1634 pKa = 3.63TLDD1637 pKa = 3.7GAVGNDD1643 pKa = 3.44TLLGGLGNDD1652 pKa = 3.45VLTGGSGADD1661 pKa = 3.45LFAFAAGDD1669 pKa = 3.9GQDD1672 pKa = 3.72TITDD1676 pKa = 4.08FDD1678 pKa = 4.0AAQGDD1683 pKa = 4.36RR1684 pKa = 11.84IGLAVGQSYY1693 pKa = 11.05SVDD1696 pKa = 3.48TNSNGDD1702 pKa = 3.37AVIVYY1707 pKa = 7.3GTSDD1711 pKa = 2.94IVTLVGIQPAAVSSTWFVTLL1731 pKa = 4.32
MM1 pKa = 7.62ASGISFLAGSTVKK14 pKa = 10.57SVNTYY19 pKa = 8.5TSSNQSYY26 pKa = 8.74PAIGSFNDD34 pKa = 3.38GGYY37 pKa = 8.4VVSWISYY44 pKa = 8.63LQDD47 pKa = 3.02GSGWGVYY54 pKa = 7.15GQRR57 pKa = 11.84FSTDD61 pKa = 4.16GIPVGGEE68 pKa = 3.26FRR70 pKa = 11.84INVSTTGDD78 pKa = 3.32QDD80 pKa = 3.63SPSIVTHH87 pKa = 6.48SDD89 pKa = 2.8GSFNVYY95 pKa = 7.51WASYY99 pKa = 7.98ATAGVPYY106 pKa = 8.13TTAANIYY113 pKa = 9.37GRR115 pKa = 11.84RR116 pKa = 11.84FDD118 pKa = 4.99ASGNPLGSEE127 pKa = 4.17FKK129 pKa = 10.98VNSATTSDD137 pKa = 3.56RR138 pKa = 11.84NRR140 pKa = 11.84VGTAKK145 pKa = 10.54LADD148 pKa = 3.97GGEE151 pKa = 4.39VVVWTLRR158 pKa = 11.84NAAWTTAEE166 pKa = 4.94LYY168 pKa = 10.5AQRR171 pKa = 11.84FDD173 pKa = 3.65SAGNSVGGEE182 pKa = 3.65FRR184 pKa = 11.84VNPTVTAASSGQPNSVAGLDD204 pKa = 3.56GGGFAVVWDD213 pKa = 4.22ASDD216 pKa = 3.97GNGSGVYY223 pKa = 7.74VQRR226 pKa = 11.84FDD228 pKa = 4.09ASGNRR233 pKa = 11.84VGMEE237 pKa = 3.66TRR239 pKa = 11.84VNSTVASDD247 pKa = 3.4QSMGNITALAGGGFVVSWTSNGQDD271 pKa = 3.07GGGTGVYY278 pKa = 9.81AQRR281 pKa = 11.84FDD283 pKa = 3.83AAGTPVGTEE292 pKa = 3.93FQVNTQTAANQYY304 pKa = 8.91GSSAIGSPDD313 pKa = 3.03GGFTILYY320 pKa = 6.96TAGDD324 pKa = 3.89NQDD327 pKa = 2.92GSGAGVYY334 pKa = 7.88GQRR337 pKa = 11.84YY338 pKa = 8.88DD339 pKa = 3.73SNGLRR344 pKa = 11.84VGGEE348 pKa = 3.63FRR350 pKa = 11.84VNDD353 pKa = 3.86STVGNQDD360 pKa = 2.86AQVIGVRR367 pKa = 11.84PDD369 pKa = 2.93GGFVVAWNSPDD380 pKa = 3.43ASQSGVLTRR389 pKa = 11.84IYY391 pKa = 10.88SNPTAPYY398 pKa = 9.89SVAGEE403 pKa = 3.9QRR405 pKa = 11.84VNTTGTGDD413 pKa = 3.4QTRR416 pKa = 11.84SSISVFADD424 pKa = 3.34GGYY427 pKa = 9.8VVTWVSAGQAGSGDD441 pKa = 3.3GVYY444 pKa = 10.46AQRR447 pKa = 11.84YY448 pKa = 8.61DD449 pKa = 3.65SGGKK453 pKa = 9.42PIGSEE458 pKa = 3.86FRR460 pKa = 11.84VNTTTTGDD468 pKa = 3.29QGDD471 pKa = 4.3ATVVAYY477 pKa = 10.37DD478 pKa = 4.03GGQFAIFWASHH489 pKa = 4.15TQAGQTSPVDD499 pKa = 3.25ASVRR503 pKa = 11.84GQRR506 pKa = 11.84YY507 pKa = 8.4AADD510 pKa = 3.77GTPVGSEE517 pKa = 3.94FQVNGASTSDD527 pKa = 3.62RR528 pKa = 11.84NPPGAVRR535 pKa = 11.84LADD538 pKa = 3.67GSTVVVWLQHH548 pKa = 5.94DD549 pKa = 4.31SSWAHH554 pKa = 4.28TWVYY558 pKa = 8.8GQRR561 pKa = 11.84YY562 pKa = 8.46DD563 pKa = 3.43AAGNAVGGEE572 pKa = 3.93FLVNSVNSANSGYY585 pKa = 9.95VASGYY590 pKa = 10.29QPASVTALEE599 pKa = 4.3NGGFAVVWNAADD611 pKa = 3.86GDD613 pKa = 4.14GTGAYY618 pKa = 8.22FQRR621 pKa = 11.84FDD623 pKa = 3.33ASANRR628 pKa = 11.84VGGEE632 pKa = 3.75VRR634 pKa = 11.84LNTTTASDD642 pKa = 3.32QFTASIAGLSGGGFVATWTSIGQDD666 pKa = 2.65GGTYY670 pKa = 10.33GIYY673 pKa = 10.38AQRR676 pKa = 11.84FDD678 pKa = 3.64SAGNAVGGEE687 pKa = 3.94FRR689 pKa = 11.84VNTITQGDD697 pKa = 3.98QYY699 pKa = 11.11ASRR702 pKa = 11.84VASLPDD708 pKa = 3.24GGFVVAYY715 pKa = 9.32SSSGGEE721 pKa = 4.11DD722 pKa = 3.3GSGIGVFGQRR732 pKa = 11.84YY733 pKa = 8.12DD734 pKa = 3.61AQGTAVGSEE743 pKa = 4.38FIITDD748 pKa = 3.58TTVGNQEE755 pKa = 4.13EE756 pKa = 4.38PHH758 pKa = 5.88LAARR762 pKa = 11.84PDD764 pKa = 3.48GGFVLTWVSPDD775 pKa = 3.0GNGRR779 pKa = 11.84GVFSRR784 pKa = 11.84LVRR787 pKa = 11.84AMAPPSATQAGTAGNDD803 pKa = 3.46LLTPANPRR811 pKa = 11.84SVLLGLAGNDD821 pKa = 3.48TLTGGSLTDD830 pKa = 3.66TLNGGDD836 pKa = 4.46GNDD839 pKa = 3.82SLSGGLGDD847 pKa = 5.22DD848 pKa = 3.92VLFGGIGNDD857 pKa = 3.5TLDD860 pKa = 3.87GGGGADD866 pKa = 4.36LLYY869 pKa = 11.08GGAGNDD875 pKa = 3.47TFIVRR880 pKa = 11.84GGEE883 pKa = 3.99ALSDD887 pKa = 3.21TGGIDD892 pKa = 3.37TAILTGPASYY902 pKa = 9.26TLPGGIEE909 pKa = 4.02NLVLTSAATGTGNSLDD925 pKa = 3.75NLLTGSSGSDD935 pKa = 3.05QLFGLAGNDD944 pKa = 3.56TLDD947 pKa = 3.8GGGGADD953 pKa = 3.67TLIGGLGNDD962 pKa = 3.89TYY964 pKa = 11.11IVDD967 pKa = 4.98DD968 pKa = 4.13IGDD971 pKa = 3.87VVTEE975 pKa = 4.23LSGEE979 pKa = 4.18GADD982 pKa = 4.04EE983 pKa = 4.19VRR985 pKa = 11.84TTLASYY991 pKa = 9.4TLGTNLEE998 pKa = 4.23NLTFTGSGSFTGTGNGLANILQGGAGNDD1026 pKa = 3.81TLDD1029 pKa = 3.78GGAGIDD1035 pKa = 3.6TLRR1038 pKa = 11.84GGVGNDD1044 pKa = 3.19TYY1046 pKa = 11.34LVSDD1050 pKa = 3.76VGDD1053 pKa = 3.84VIIEE1057 pKa = 4.1AVGEE1061 pKa = 4.3GTDD1064 pKa = 3.72EE1065 pKa = 4.28VRR1067 pKa = 11.84TTLASYY1073 pKa = 9.03TLAANVEE1080 pKa = 4.23NLVYY1084 pKa = 10.02TGTVAFTGTGNTGNNAITGGAGNDD1108 pKa = 3.95SLTGDD1113 pKa = 4.3AGNDD1117 pKa = 3.69TLDD1120 pKa = 3.9GGAGADD1126 pKa = 3.6TLVGGTGNDD1135 pKa = 3.43TYY1137 pKa = 11.11IVDD1140 pKa = 3.72TAGDD1144 pKa = 4.07VVTEE1148 pKa = 4.23LAGQGTDD1155 pKa = 3.15TVRR1158 pKa = 11.84TSLASYY1164 pKa = 9.13TLGANVEE1171 pKa = 4.28NLVFTGSGPVTFTGNALNNNLTGGAGADD1199 pKa = 4.07LLTGLDD1205 pKa = 4.67GNDD1208 pKa = 3.75TLNGGAGADD1217 pKa = 3.71TLLGGTGNDD1226 pKa = 3.28TYY1228 pKa = 11.76VVDD1231 pKa = 3.75NVGDD1235 pKa = 3.89VLVEE1239 pKa = 3.98LPGEE1243 pKa = 4.65GIDD1246 pKa = 3.89TVQSSINHH1254 pKa = 5.77TLLGDD1259 pKa = 3.98FEE1261 pKa = 5.41NLTLTGSAALTGTGNALNNLLTGNGAANLLTGLEE1295 pKa = 4.45GDD1297 pKa = 3.88DD1298 pKa = 3.88TLNGGGGADD1307 pKa = 3.62TLIGGLGNDD1316 pKa = 3.84SYY1318 pKa = 12.11VVDD1321 pKa = 3.72NAGDD1325 pKa = 3.74VVTEE1329 pKa = 3.96LVGEE1333 pKa = 4.83GIDD1336 pKa = 3.63TVNASINWTLGTNLEE1351 pKa = 4.13NLTLTGSAAISATGNGLNNLLTGNSGANLLDD1382 pKa = 4.28GGLGADD1388 pKa = 4.05SMVGGAGNDD1397 pKa = 3.43VYY1399 pKa = 11.0IVDD1402 pKa = 4.56DD1403 pKa = 4.64ADD1405 pKa = 4.04DD1406 pKa = 3.92VVTEE1410 pKa = 4.42LVGQGTDD1417 pKa = 3.14EE1418 pKa = 3.99VRR1420 pKa = 11.84TNLSSYY1426 pKa = 11.33LLGTNIEE1433 pKa = 4.44ALTHH1437 pKa = 6.16TGTGDD1442 pKa = 3.36FAGTGNALDD1451 pKa = 3.89NLLTGGLGNDD1461 pKa = 3.86TLSGGDD1467 pKa = 3.84GADD1470 pKa = 3.71TLDD1473 pKa = 4.34GGAGSNLLIGGTGHH1487 pKa = 7.61DD1488 pKa = 3.83VYY1490 pKa = 11.35LVGGFGDD1497 pKa = 3.54QVVEE1501 pKa = 3.93LVGEE1505 pKa = 4.35GMDD1508 pKa = 3.63EE1509 pKa = 4.06VRR1511 pKa = 11.84TALSGYY1517 pKa = 7.4TLTDD1521 pKa = 3.37GVEE1524 pKa = 4.1TLTYY1528 pKa = 9.09TGVAAFVGTGNGLDD1542 pKa = 3.56NLIAGGAGADD1552 pKa = 3.82TLDD1555 pKa = 4.19GGLGADD1561 pKa = 3.88TLIGGLGDD1569 pKa = 3.6DD1570 pKa = 4.24VYY1572 pKa = 11.74VVDD1575 pKa = 4.37SAGDD1579 pKa = 3.77TVIEE1583 pKa = 4.42GADD1586 pKa = 3.23AGLDD1590 pKa = 3.6EE1591 pKa = 5.1VRR1593 pKa = 11.84TTLSAFALGDD1603 pKa = 3.34NTEE1606 pKa = 4.05RR1607 pKa = 11.84LVFQGSGDD1615 pKa = 3.32ASGIGNGLDD1624 pKa = 2.81NWLAGNIGNDD1634 pKa = 3.63TLDD1637 pKa = 3.7GAVGNDD1643 pKa = 3.44TLLGGLGNDD1652 pKa = 3.45VLTGGSGADD1661 pKa = 3.45LFAFAAGDD1669 pKa = 3.9GQDD1672 pKa = 3.72TITDD1676 pKa = 4.08FDD1678 pKa = 4.0AAQGDD1683 pKa = 4.36RR1684 pKa = 11.84IGLAVGQSYY1693 pKa = 11.05SVDD1696 pKa = 3.48TNSNGDD1702 pKa = 3.37AVIVYY1707 pKa = 7.3GTSDD1711 pKa = 2.94IVTLVGIQPAAVSSTWFVTLL1731 pKa = 4.32
Molecular weight: 172.92 kDa
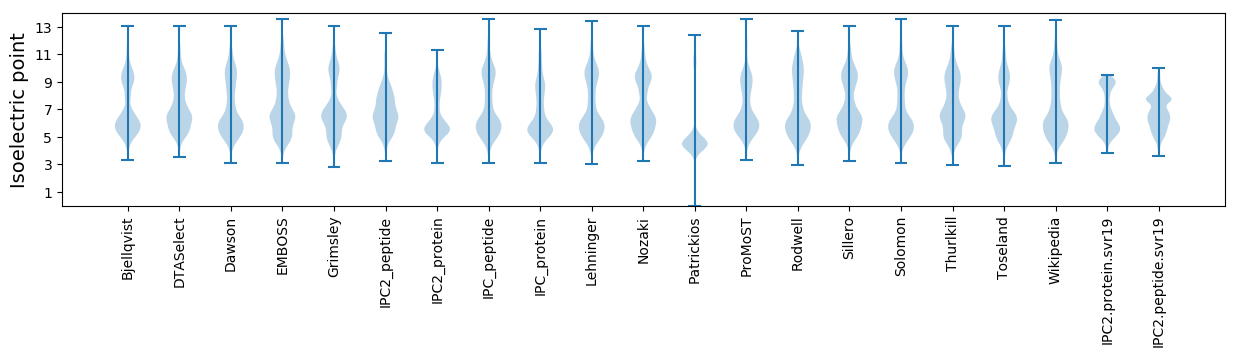
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A431VHE3|A0A431VHE3_9PROT Propionyl-CoA synthetase OS=Azospirillum sp. L-25-5w-1 OX=2496639 GN=EJ903_10570 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 8.96IVRR12 pKa = 11.84KK13 pKa = 9.24RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.3VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.91VLSAA44 pKa = 4.11
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 8.96IVRR12 pKa = 11.84KK13 pKa = 9.24RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.3VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.91VLSAA44 pKa = 4.11
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1727630 |
29 |
5758 |
334.8 |
36.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.313 ± 0.055 | 0.859 ± 0.01 |
5.992 ± 0.026 | 5.027 ± 0.032 |
3.359 ± 0.024 | 8.694 ± 0.042 |
2.077 ± 0.016 | 4.462 ± 0.026 |
2.708 ± 0.028 | 10.82 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.265 ± 0.015 | 2.424 ± 0.019 |
5.547 ± 0.035 | 3.012 ± 0.02 |
7.66 ± 0.039 | 5.128 ± 0.028 |
5.66 ± 0.033 | 7.758 ± 0.026 |
1.298 ± 0.014 | 1.939 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
