
Wuhan arthropod virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.47
Get precalculated fractions of proteins
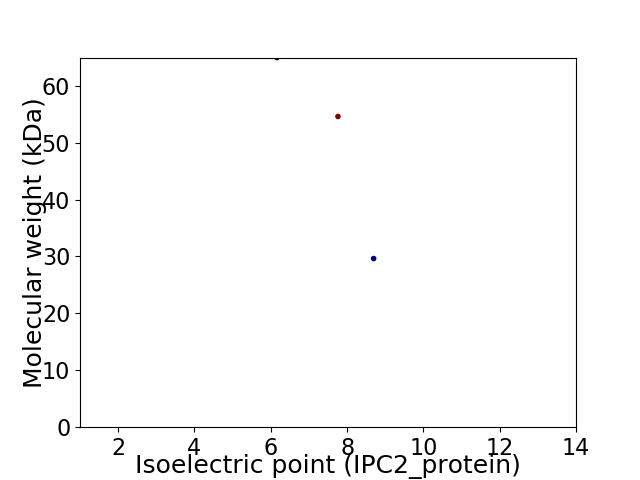
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KF51|A0A1L3KF51_9VIRU Uncharacterized protein OS=Wuhan arthropod virus 4 OX=1923693 PE=4 SV=1
MM1 pKa = 7.48SAEE4 pKa = 4.0QVQTPVNFVLQAPAGAFAAPPPAEE28 pKa = 4.23SHH30 pKa = 5.19WLVRR34 pKa = 11.84AYY36 pKa = 9.79KK37 pKa = 10.31RR38 pKa = 11.84LGSLWPLFLASWWFSIPNLMQVLVGAYY65 pKa = 9.36FDD67 pKa = 3.74KK68 pKa = 11.24LGWLVDD74 pKa = 3.87LYY76 pKa = 11.69GRR78 pKa = 11.84VEE80 pKa = 3.87YY81 pKa = 10.89SADD84 pKa = 3.13KK85 pKa = 10.62GIQYY89 pKa = 10.18VVDD92 pKa = 3.54YY93 pKa = 10.08RR94 pKa = 11.84LPLILGTCTTLIWACATAMMWKK116 pKa = 9.33WMNDD120 pKa = 3.44DD121 pKa = 3.69VKK123 pKa = 10.97DD124 pKa = 3.84TIDD127 pKa = 4.11RR128 pKa = 11.84EE129 pKa = 4.45VQVRR133 pKa = 11.84LLQEE137 pKa = 4.27GLAKK141 pKa = 10.1QAATQTVTDD150 pKa = 4.15PVEE153 pKa = 4.01FTPEE157 pKa = 3.74KK158 pKa = 10.33VKK160 pKa = 10.67PSSILIKK167 pKa = 9.72PEE169 pKa = 3.78RR170 pKa = 11.84PTFQAAIYY178 pKa = 10.63GEE180 pKa = 4.96LNGAWVRR187 pKa = 11.84SGQAFKK193 pKa = 10.97VRR195 pKa = 11.84HH196 pKa = 4.69QLITAYY202 pKa = 10.09HH203 pKa = 5.03VVEE206 pKa = 4.18SFQRR210 pKa = 11.84LRR212 pKa = 11.84LVTSSGEE219 pKa = 3.2IDD221 pKa = 3.29MASEE225 pKa = 4.28RR226 pKa = 11.84FRR228 pKa = 11.84WVEE231 pKa = 3.65GDD233 pKa = 3.68VAFCLLTPQEE243 pKa = 4.14NQILALTEE251 pKa = 4.03AKK253 pKa = 10.15LANHH257 pKa = 6.57EE258 pKa = 4.19FASNSGLIVKK268 pKa = 8.38ATGFNNSSYY277 pKa = 11.14GFLKK281 pKa = 10.22EE282 pKa = 4.05YY283 pKa = 10.09PAFGYY288 pKa = 10.23VEE290 pKa = 4.4YY291 pKa = 10.74AGSTVPGFSGCPYY304 pKa = 9.39VVGPKK309 pKa = 10.08VYY311 pKa = 10.27GMHH314 pKa = 7.11LGGGVEE320 pKa = 4.03NLGLNGAYY328 pKa = 10.46LDD330 pKa = 3.56MLVRR334 pKa = 11.84RR335 pKa = 11.84KK336 pKa = 10.42DD337 pKa = 3.65EE338 pKa = 3.96SSEE341 pKa = 4.31DD342 pKa = 3.59YY343 pKa = 11.29LFGQIEE349 pKa = 4.08KK350 pKa = 10.31HH351 pKa = 5.7EE352 pKa = 4.65KK353 pKa = 10.04YY354 pKa = 10.4DD355 pKa = 4.05YY356 pKa = 10.46QQSPYY361 pKa = 11.09DD362 pKa = 3.67PDD364 pKa = 3.45EE365 pKa = 4.05FRR367 pKa = 11.84LRR369 pKa = 11.84IGGKK373 pKa = 10.13YY374 pKa = 9.87YY375 pKa = 10.86LIDD378 pKa = 4.59ADD380 pKa = 4.17MLHH383 pKa = 7.15SLEE386 pKa = 4.28TRR388 pKa = 11.84QKK390 pKa = 9.67GAGSNKK396 pKa = 9.6VMFKK400 pKa = 10.6DD401 pKa = 4.01LEE403 pKa = 4.32YY404 pKa = 10.91DD405 pKa = 3.39AEE407 pKa = 4.77AYY409 pKa = 7.91EE410 pKa = 4.28YY411 pKa = 8.51PTEE414 pKa = 4.2EE415 pKa = 3.67EE416 pKa = 4.76HH417 pKa = 7.56IPVYY421 pKa = 10.11KK422 pKa = 10.27PKK424 pKa = 10.1EE425 pKa = 3.7IPKK428 pKa = 9.8IDD430 pKa = 3.88DD431 pKa = 4.31EE432 pKa = 4.35IGKK435 pKa = 9.68VSKK438 pKa = 10.58EE439 pKa = 3.36IAGLIKK445 pKa = 10.31EE446 pKa = 3.95ISEE449 pKa = 3.9NRR451 pKa = 11.84NAPTASKK458 pKa = 10.11PVGAAGPRR466 pKa = 11.84KK467 pKa = 9.75QPVSAQKK474 pKa = 10.35QAVARR479 pKa = 11.84PKK481 pKa = 9.85GTTSSSPKK489 pKa = 10.06VSVTDD494 pKa = 4.28GPEE497 pKa = 3.71LTPVQRR503 pKa = 11.84SQVLQDD509 pKa = 3.12IAKK512 pKa = 9.98SLRR515 pKa = 11.84TLRR518 pKa = 11.84AEE520 pKa = 4.39QPRR523 pKa = 11.84LQQAISKK530 pKa = 9.55KK531 pKa = 9.55SSKK534 pKa = 10.48SPQAASQIGRR544 pKa = 11.84LISLNTEE551 pKa = 3.86LQRR554 pKa = 11.84IISEE558 pKa = 4.4FGLMTPLPTSTEE570 pKa = 3.52TRR572 pKa = 11.84ARR574 pKa = 11.84DD575 pKa = 3.51VASTSVSPPP584 pKa = 3.05
MM1 pKa = 7.48SAEE4 pKa = 4.0QVQTPVNFVLQAPAGAFAAPPPAEE28 pKa = 4.23SHH30 pKa = 5.19WLVRR34 pKa = 11.84AYY36 pKa = 9.79KK37 pKa = 10.31RR38 pKa = 11.84LGSLWPLFLASWWFSIPNLMQVLVGAYY65 pKa = 9.36FDD67 pKa = 3.74KK68 pKa = 11.24LGWLVDD74 pKa = 3.87LYY76 pKa = 11.69GRR78 pKa = 11.84VEE80 pKa = 3.87YY81 pKa = 10.89SADD84 pKa = 3.13KK85 pKa = 10.62GIQYY89 pKa = 10.18VVDD92 pKa = 3.54YY93 pKa = 10.08RR94 pKa = 11.84LPLILGTCTTLIWACATAMMWKK116 pKa = 9.33WMNDD120 pKa = 3.44DD121 pKa = 3.69VKK123 pKa = 10.97DD124 pKa = 3.84TIDD127 pKa = 4.11RR128 pKa = 11.84EE129 pKa = 4.45VQVRR133 pKa = 11.84LLQEE137 pKa = 4.27GLAKK141 pKa = 10.1QAATQTVTDD150 pKa = 4.15PVEE153 pKa = 4.01FTPEE157 pKa = 3.74KK158 pKa = 10.33VKK160 pKa = 10.67PSSILIKK167 pKa = 9.72PEE169 pKa = 3.78RR170 pKa = 11.84PTFQAAIYY178 pKa = 10.63GEE180 pKa = 4.96LNGAWVRR187 pKa = 11.84SGQAFKK193 pKa = 10.97VRR195 pKa = 11.84HH196 pKa = 4.69QLITAYY202 pKa = 10.09HH203 pKa = 5.03VVEE206 pKa = 4.18SFQRR210 pKa = 11.84LRR212 pKa = 11.84LVTSSGEE219 pKa = 3.2IDD221 pKa = 3.29MASEE225 pKa = 4.28RR226 pKa = 11.84FRR228 pKa = 11.84WVEE231 pKa = 3.65GDD233 pKa = 3.68VAFCLLTPQEE243 pKa = 4.14NQILALTEE251 pKa = 4.03AKK253 pKa = 10.15LANHH257 pKa = 6.57EE258 pKa = 4.19FASNSGLIVKK268 pKa = 8.38ATGFNNSSYY277 pKa = 11.14GFLKK281 pKa = 10.22EE282 pKa = 4.05YY283 pKa = 10.09PAFGYY288 pKa = 10.23VEE290 pKa = 4.4YY291 pKa = 10.74AGSTVPGFSGCPYY304 pKa = 9.39VVGPKK309 pKa = 10.08VYY311 pKa = 10.27GMHH314 pKa = 7.11LGGGVEE320 pKa = 4.03NLGLNGAYY328 pKa = 10.46LDD330 pKa = 3.56MLVRR334 pKa = 11.84RR335 pKa = 11.84KK336 pKa = 10.42DD337 pKa = 3.65EE338 pKa = 3.96SSEE341 pKa = 4.31DD342 pKa = 3.59YY343 pKa = 11.29LFGQIEE349 pKa = 4.08KK350 pKa = 10.31HH351 pKa = 5.7EE352 pKa = 4.65KK353 pKa = 10.04YY354 pKa = 10.4DD355 pKa = 4.05YY356 pKa = 10.46QQSPYY361 pKa = 11.09DD362 pKa = 3.67PDD364 pKa = 3.45EE365 pKa = 4.05FRR367 pKa = 11.84LRR369 pKa = 11.84IGGKK373 pKa = 10.13YY374 pKa = 9.87YY375 pKa = 10.86LIDD378 pKa = 4.59ADD380 pKa = 4.17MLHH383 pKa = 7.15SLEE386 pKa = 4.28TRR388 pKa = 11.84QKK390 pKa = 9.67GAGSNKK396 pKa = 9.6VMFKK400 pKa = 10.6DD401 pKa = 4.01LEE403 pKa = 4.32YY404 pKa = 10.91DD405 pKa = 3.39AEE407 pKa = 4.77AYY409 pKa = 7.91EE410 pKa = 4.28YY411 pKa = 8.51PTEE414 pKa = 4.2EE415 pKa = 3.67EE416 pKa = 4.76HH417 pKa = 7.56IPVYY421 pKa = 10.11KK422 pKa = 10.27PKK424 pKa = 10.1EE425 pKa = 3.7IPKK428 pKa = 9.8IDD430 pKa = 3.88DD431 pKa = 4.31EE432 pKa = 4.35IGKK435 pKa = 9.68VSKK438 pKa = 10.58EE439 pKa = 3.36IAGLIKK445 pKa = 10.31EE446 pKa = 3.95ISEE449 pKa = 3.9NRR451 pKa = 11.84NAPTASKK458 pKa = 10.11PVGAAGPRR466 pKa = 11.84KK467 pKa = 9.75QPVSAQKK474 pKa = 10.35QAVARR479 pKa = 11.84PKK481 pKa = 9.85GTTSSSPKK489 pKa = 10.06VSVTDD494 pKa = 4.28GPEE497 pKa = 3.71LTPVQRR503 pKa = 11.84SQVLQDD509 pKa = 3.12IAKK512 pKa = 9.98SLRR515 pKa = 11.84TLRR518 pKa = 11.84AEE520 pKa = 4.39QPRR523 pKa = 11.84LQQAISKK530 pKa = 9.55KK531 pKa = 9.55SSKK534 pKa = 10.48SPQAASQIGRR544 pKa = 11.84LISLNTEE551 pKa = 3.86LQRR554 pKa = 11.84IISEE558 pKa = 4.4FGLMTPLPTSTEE570 pKa = 3.52TRR572 pKa = 11.84ARR574 pKa = 11.84DD575 pKa = 3.51VASTSVSPPP584 pKa = 3.05
Molecular weight: 64.93 kDa
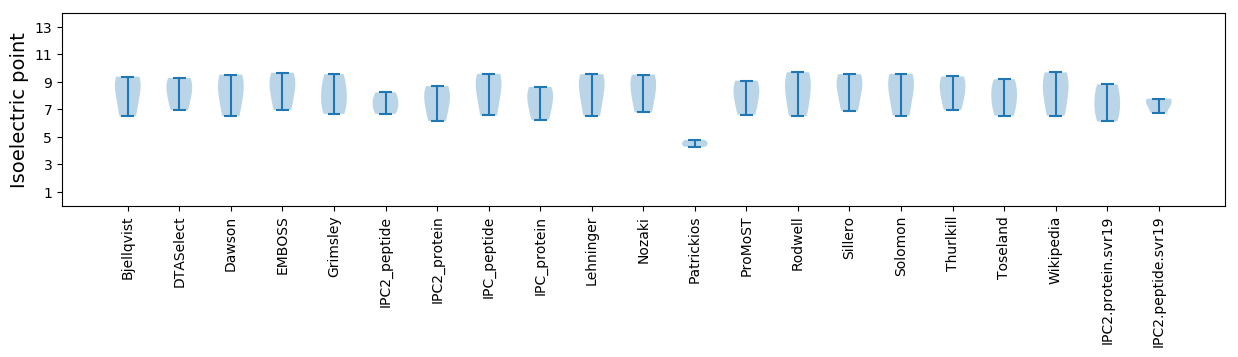
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEY9|A0A1L3KEY9_9VIRU Capsid protein OS=Wuhan arthropod virus 4 OX=1923693 PE=3 SV=1
MM1 pKa = 7.48VDD3 pKa = 3.03RR4 pKa = 11.84VLFGWLAQAALSNVGKK20 pKa = 8.23TPCAVGWAPVKK31 pKa = 10.73SGWRR35 pKa = 11.84VVRR38 pKa = 11.84HH39 pKa = 6.08RR40 pKa = 11.84FPKK43 pKa = 10.25SICLDD48 pKa = 3.14KK49 pKa = 11.27SAWDD53 pKa = 3.38WTVQPWLITAYY64 pKa = 10.19HH65 pKa = 6.28RR66 pKa = 11.84FVNEE70 pKa = 4.05LALEE74 pKa = 4.37APDD77 pKa = 3.08WWRR80 pKa = 11.84KK81 pKa = 8.47AVHH84 pKa = 6.35SRR86 pKa = 11.84FTLLFRR92 pKa = 11.84AAKK95 pKa = 9.22FQFRR99 pKa = 11.84NGTVIAQPEE108 pKa = 4.37VGVMKK113 pKa = 10.38SGCYY117 pKa = 8.49LTLLLNSVGQVLAHH131 pKa = 6.76IAAKK135 pKa = 10.3LRR137 pKa = 11.84LGEE140 pKa = 4.7DD141 pKa = 3.98PYY143 pKa = 11.59EE144 pKa = 4.31SLPFAMGDD152 pKa = 3.47DD153 pKa = 4.26TVQSPRR159 pKa = 11.84GIHH162 pKa = 6.6DD163 pKa = 3.65LEE165 pKa = 4.94SYY167 pKa = 10.54VRR169 pKa = 11.84EE170 pKa = 4.18LEE172 pKa = 4.25KK173 pKa = 10.95LGCKK177 pKa = 9.95VKK179 pKa = 10.65GAVVRR184 pKa = 11.84PDD186 pKa = 3.09VEE188 pKa = 4.06FAGFQMTDD196 pKa = 3.15RR197 pKa = 11.84RR198 pKa = 11.84CVPAYY203 pKa = 10.0RR204 pKa = 11.84KK205 pKa = 9.24KK206 pKa = 9.97HH207 pKa = 5.33LYY209 pKa = 8.98KK210 pKa = 10.63LEE212 pKa = 4.11YY213 pKa = 9.81ATDD216 pKa = 3.79LPAYY220 pKa = 9.05LRR222 pKa = 11.84SMQLLYY228 pKa = 11.13GHH230 pKa = 7.17DD231 pKa = 3.14HH232 pKa = 5.93TMFSFFHH239 pKa = 5.86NVAARR244 pKa = 11.84KK245 pKa = 9.53CPEE248 pKa = 3.78AAISTSTAMMFMGG261 pKa = 4.98
MM1 pKa = 7.48VDD3 pKa = 3.03RR4 pKa = 11.84VLFGWLAQAALSNVGKK20 pKa = 8.23TPCAVGWAPVKK31 pKa = 10.73SGWRR35 pKa = 11.84VVRR38 pKa = 11.84HH39 pKa = 6.08RR40 pKa = 11.84FPKK43 pKa = 10.25SICLDD48 pKa = 3.14KK49 pKa = 11.27SAWDD53 pKa = 3.38WTVQPWLITAYY64 pKa = 10.19HH65 pKa = 6.28RR66 pKa = 11.84FVNEE70 pKa = 4.05LALEE74 pKa = 4.37APDD77 pKa = 3.08WWRR80 pKa = 11.84KK81 pKa = 8.47AVHH84 pKa = 6.35SRR86 pKa = 11.84FTLLFRR92 pKa = 11.84AAKK95 pKa = 9.22FQFRR99 pKa = 11.84NGTVIAQPEE108 pKa = 4.37VGVMKK113 pKa = 10.38SGCYY117 pKa = 8.49LTLLLNSVGQVLAHH131 pKa = 6.76IAAKK135 pKa = 10.3LRR137 pKa = 11.84LGEE140 pKa = 4.7DD141 pKa = 3.98PYY143 pKa = 11.59EE144 pKa = 4.31SLPFAMGDD152 pKa = 3.47DD153 pKa = 4.26TVQSPRR159 pKa = 11.84GIHH162 pKa = 6.6DD163 pKa = 3.65LEE165 pKa = 4.94SYY167 pKa = 10.54VRR169 pKa = 11.84EE170 pKa = 4.18LEE172 pKa = 4.25KK173 pKa = 10.95LGCKK177 pKa = 9.95VKK179 pKa = 10.65GAVVRR184 pKa = 11.84PDD186 pKa = 3.09VEE188 pKa = 4.06FAGFQMTDD196 pKa = 3.15RR197 pKa = 11.84RR198 pKa = 11.84CVPAYY203 pKa = 10.0RR204 pKa = 11.84KK205 pKa = 9.24KK206 pKa = 9.97HH207 pKa = 5.33LYY209 pKa = 8.98KK210 pKa = 10.63LEE212 pKa = 4.11YY213 pKa = 9.81ATDD216 pKa = 3.79LPAYY220 pKa = 9.05LRR222 pKa = 11.84SMQLLYY228 pKa = 11.13GHH230 pKa = 7.17DD231 pKa = 3.14HH232 pKa = 5.93TMFSFFHH239 pKa = 5.86NVAARR244 pKa = 11.84KK245 pKa = 9.53CPEE248 pKa = 3.78AAISTSTAMMFMGG261 pKa = 4.98
Molecular weight: 29.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1332 |
261 |
584 |
444.0 |
49.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.859 ± 0.386 | 0.976 ± 0.328 |
4.73 ± 0.181 | 5.631 ± 0.72 |
3.529 ± 0.486 | 5.931 ± 0.56 |
2.628 ± 0.659 | 4.129 ± 0.48 |
5.706 ± 0.272 | 9.61 ± 0.375 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.177 ± 0.315 | 2.778 ± 0.314 |
6.757 ± 0.574 | 4.129 ± 0.61 |
5.48 ± 0.371 | 6.832 ± 0.486 |
6.381 ± 0.956 | 7.508 ± 0.333 |
2.402 ± 0.357 | 3.754 ± 0.278 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
