
Hubei tombus-like virus 29
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.6
Get precalculated fractions of proteins
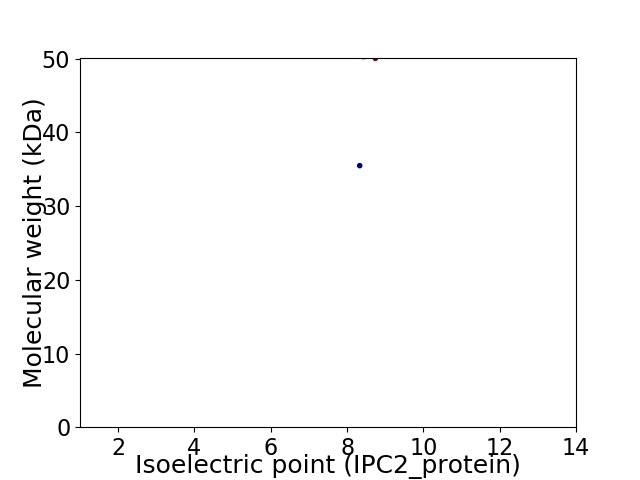
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGC0|A0A1L3KGC0_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 29 OX=1923276 PE=4 SV=1
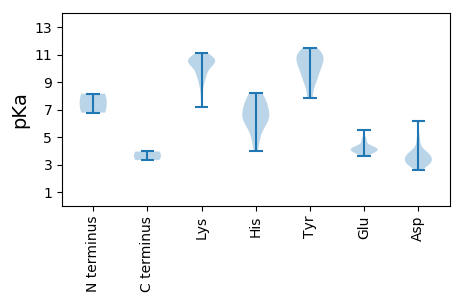
MM1 pKa = 8.15RR2 pKa = 11.84PRR4 pKa = 11.84LVMNRR9 pKa = 11.84SVSDD13 pKa = 3.52PAFVSWVIGDD23 pKa = 4.04PLLSNRR29 pKa = 11.84EE30 pKa = 3.75ARR32 pKa = 11.84NGPKK36 pKa = 9.73RR37 pKa = 11.84HH38 pKa = 6.29SDD40 pKa = 3.08SSIGPTRR47 pKa = 11.84QPAYY51 pKa = 9.65VTTLHH56 pKa = 6.19NVAAEE61 pKa = 4.29TFGKK65 pKa = 9.44LLSMGYY71 pKa = 9.91DD72 pKa = 3.4VQWNYY77 pKa = 10.32ICNLILQHH85 pKa = 6.79GIGSTEE91 pKa = 4.07RR92 pKa = 11.84IIDD95 pKa = 3.39IARR98 pKa = 11.84NEE100 pKa = 4.14CSMIPKK106 pKa = 10.32FSDD109 pKa = 3.16NEE111 pKa = 4.12GRR113 pKa = 11.84QHH115 pKa = 7.49LEE117 pKa = 3.87AVDD120 pKa = 3.55KK121 pKa = 10.77HH122 pKa = 6.37DD123 pKa = 6.18KK124 pKa = 10.48IVADD128 pKa = 3.79TKK130 pKa = 10.69LVKK133 pKa = 10.7AEE135 pKa = 3.71ARR137 pKa = 11.84KK138 pKa = 9.81IEE140 pKa = 4.07LEE142 pKa = 3.92NVKK145 pKa = 10.54LEE147 pKa = 4.17KK148 pKa = 10.96EE149 pKa = 4.1NVLLEE154 pKa = 4.29KK155 pKa = 10.91GLDD158 pKa = 3.38IKK160 pKa = 11.03KK161 pKa = 10.39LEE163 pKa = 4.05QTIQQLGMEE172 pKa = 4.59KK173 pKa = 10.07EE174 pKa = 3.99HH175 pKa = 7.69AILLKK180 pKa = 10.6NDD182 pKa = 2.93IDD184 pKa = 3.69ILDD187 pKa = 3.97AYY189 pKa = 10.53GYY191 pKa = 10.91LCAQTIGQQITNKK204 pKa = 10.25HH205 pKa = 4.83EE206 pKa = 4.24TTCANICRR214 pKa = 11.84NYY216 pKa = 9.61FAKK219 pKa = 10.63SQITDD224 pKa = 2.61NMMILHH230 pKa = 8.15AIRR233 pKa = 11.84IILAMYY239 pKa = 10.26SVDD242 pKa = 3.7RR243 pKa = 11.84KK244 pKa = 10.2FAMTSYY250 pKa = 10.84FDD252 pKa = 3.52WDD254 pKa = 3.62AKK256 pKa = 11.11GKK258 pKa = 10.46DD259 pKa = 3.64DD260 pKa = 5.09FEE262 pKa = 4.84KK263 pKa = 10.7LRR265 pKa = 11.84KK266 pKa = 9.36KK267 pKa = 10.79ADD269 pKa = 3.55VEE271 pKa = 4.25HH272 pKa = 6.87KK273 pKa = 8.46PWKK276 pKa = 8.79VIKK279 pKa = 10.69GITAVNKK286 pKa = 8.54MRR288 pKa = 11.84AGITTRR294 pKa = 11.84SRR296 pKa = 11.84FFGLFNKK303 pKa = 10.13KK304 pKa = 10.37YY305 pKa = 9.72EE306 pKa = 4.19LTSFF310 pKa = 3.95
MM1 pKa = 8.15RR2 pKa = 11.84PRR4 pKa = 11.84LVMNRR9 pKa = 11.84SVSDD13 pKa = 3.52PAFVSWVIGDD23 pKa = 4.04PLLSNRR29 pKa = 11.84EE30 pKa = 3.75ARR32 pKa = 11.84NGPKK36 pKa = 9.73RR37 pKa = 11.84HH38 pKa = 6.29SDD40 pKa = 3.08SSIGPTRR47 pKa = 11.84QPAYY51 pKa = 9.65VTTLHH56 pKa = 6.19NVAAEE61 pKa = 4.29TFGKK65 pKa = 9.44LLSMGYY71 pKa = 9.91DD72 pKa = 3.4VQWNYY77 pKa = 10.32ICNLILQHH85 pKa = 6.79GIGSTEE91 pKa = 4.07RR92 pKa = 11.84IIDD95 pKa = 3.39IARR98 pKa = 11.84NEE100 pKa = 4.14CSMIPKK106 pKa = 10.32FSDD109 pKa = 3.16NEE111 pKa = 4.12GRR113 pKa = 11.84QHH115 pKa = 7.49LEE117 pKa = 3.87AVDD120 pKa = 3.55KK121 pKa = 10.77HH122 pKa = 6.37DD123 pKa = 6.18KK124 pKa = 10.48IVADD128 pKa = 3.79TKK130 pKa = 10.69LVKK133 pKa = 10.7AEE135 pKa = 3.71ARR137 pKa = 11.84KK138 pKa = 9.81IEE140 pKa = 4.07LEE142 pKa = 3.92NVKK145 pKa = 10.54LEE147 pKa = 4.17KK148 pKa = 10.96EE149 pKa = 4.1NVLLEE154 pKa = 4.29KK155 pKa = 10.91GLDD158 pKa = 3.38IKK160 pKa = 11.03KK161 pKa = 10.39LEE163 pKa = 4.05QTIQQLGMEE172 pKa = 4.59KK173 pKa = 10.07EE174 pKa = 3.99HH175 pKa = 7.69AILLKK180 pKa = 10.6NDD182 pKa = 2.93IDD184 pKa = 3.69ILDD187 pKa = 3.97AYY189 pKa = 10.53GYY191 pKa = 10.91LCAQTIGQQITNKK204 pKa = 10.25HH205 pKa = 4.83EE206 pKa = 4.24TTCANICRR214 pKa = 11.84NYY216 pKa = 9.61FAKK219 pKa = 10.63SQITDD224 pKa = 2.61NMMILHH230 pKa = 8.15AIRR233 pKa = 11.84IILAMYY239 pKa = 10.26SVDD242 pKa = 3.7RR243 pKa = 11.84KK244 pKa = 10.2FAMTSYY250 pKa = 10.84FDD252 pKa = 3.52WDD254 pKa = 3.62AKK256 pKa = 11.11GKK258 pKa = 10.46DD259 pKa = 3.64DD260 pKa = 5.09FEE262 pKa = 4.84KK263 pKa = 10.7LRR265 pKa = 11.84KK266 pKa = 9.36KK267 pKa = 10.79ADD269 pKa = 3.55VEE271 pKa = 4.25HH272 pKa = 6.87KK273 pKa = 8.46PWKK276 pKa = 8.79VIKK279 pKa = 10.69GITAVNKK286 pKa = 8.54MRR288 pKa = 11.84AGITTRR294 pKa = 11.84SRR296 pKa = 11.84FFGLFNKK303 pKa = 10.13KK304 pKa = 10.37YY305 pKa = 9.72EE306 pKa = 4.19LTSFF310 pKa = 3.95
Molecular weight: 35.49 kDa
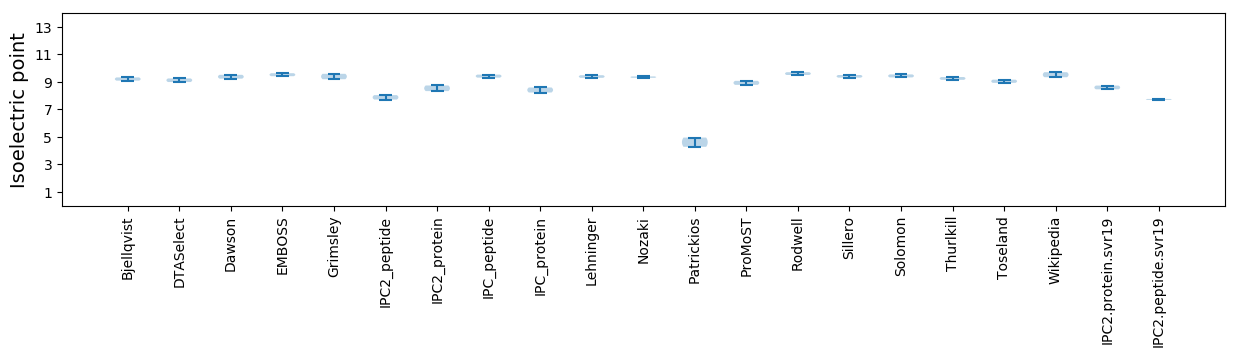
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGC0|A0A1L3KGC0_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 29 OX=1923276 PE=4 SV=1
MM1 pKa = 6.78NTQLFKK7 pKa = 10.85YY8 pKa = 9.41IKK10 pKa = 9.21KK11 pKa = 9.71FVVKK15 pKa = 10.03VEE17 pKa = 3.64PWTFDD22 pKa = 2.97EE23 pKa = 5.49YY24 pKa = 10.55IASMSSASKK33 pKa = 9.85RR34 pKa = 11.84KK35 pKa = 9.55QYY37 pKa = 11.17SDD39 pKa = 4.97YY40 pKa = 10.82YY41 pKa = 10.97DD42 pKa = 3.07QYY44 pKa = 11.14LRR46 pKa = 11.84NGRR49 pKa = 11.84IPSYY53 pKa = 9.17ITPFTKK59 pKa = 9.42IEE61 pKa = 4.01KK62 pKa = 9.28MSSGKK67 pKa = 10.38YY68 pKa = 8.78KK69 pKa = 10.37APRR72 pKa = 11.84MIQGRR77 pKa = 11.84HH78 pKa = 3.96MVFNLHH84 pKa = 5.66YY85 pKa = 10.28GRR87 pKa = 11.84YY88 pKa = 8.72IKK90 pKa = 10.31PLEE93 pKa = 4.3KK94 pKa = 10.42EE95 pKa = 3.95VTKK98 pKa = 10.79YY99 pKa = 11.08GKK101 pKa = 10.42FSIHH105 pKa = 6.35FGKK108 pKa = 10.89GNYY111 pKa = 7.84NQQAEE116 pKa = 4.8KK117 pKa = 10.42IFKK120 pKa = 10.37LSQKK124 pKa = 9.18YY125 pKa = 9.66KK126 pKa = 10.38YY127 pKa = 9.11YY128 pKa = 10.08TEE130 pKa = 4.94CDD132 pKa = 3.17HH133 pKa = 6.94TSFDD137 pKa = 3.14AHH139 pKa = 5.04VTVEE143 pKa = 4.03QLRR146 pKa = 11.84LTHH149 pKa = 6.96RR150 pKa = 11.84FYY152 pKa = 11.49NSCFPGYY159 pKa = 10.5ASDD162 pKa = 3.78VARR165 pKa = 11.84LAKK168 pKa = 9.5RR169 pKa = 11.84TINNTCISRR178 pKa = 11.84TGDD181 pKa = 2.77IYY183 pKa = 10.73RR184 pKa = 11.84VRR186 pKa = 11.84GSRR189 pKa = 11.84MSGDD193 pKa = 3.15VDD195 pKa = 3.54TGFGNCLINYY205 pKa = 8.01AILKK209 pKa = 8.09AACSNLHH216 pKa = 6.23IKK218 pKa = 10.79CEE220 pKa = 4.17IIVNGDD226 pKa = 3.26DD227 pKa = 4.55SIIFTDD233 pKa = 3.91VPIPTAEE240 pKa = 3.95LSAEE244 pKa = 3.73LRR246 pKa = 11.84KK247 pKa = 10.53YY248 pKa = 11.2NMEE251 pKa = 4.3SKK253 pKa = 10.72VNPSVSDD260 pKa = 2.93IHH262 pKa = 8.17KK263 pKa = 10.75VEE265 pKa = 4.73FCRR268 pKa = 11.84TKK270 pKa = 10.93LVFNASAKK278 pKa = 7.2PTMMIDD284 pKa = 3.68PKK286 pKa = 10.77RR287 pKa = 11.84LIDD290 pKa = 3.89IYY292 pKa = 10.83GMCYY296 pKa = 10.0TISKK300 pKa = 9.92KK301 pKa = 10.43NYY303 pKa = 7.91HH304 pKa = 6.0QYY306 pKa = 11.27LLEE309 pKa = 4.01TAMCNSYY316 pKa = 10.77INCNNPLGVLWAKK329 pKa = 9.96YY330 pKa = 10.11FNIDD334 pKa = 4.39LISYY338 pKa = 8.91KK339 pKa = 10.57KK340 pKa = 10.36NEE342 pKa = 3.77KK343 pKa = 10.54SKK345 pKa = 10.97IKK347 pKa = 10.0ILQNIKK353 pKa = 10.56CLEE356 pKa = 3.84KK357 pKa = 11.06DD358 pKa = 3.93KK359 pKa = 10.64ILKK362 pKa = 9.31MMSLSPDD369 pKa = 3.02EE370 pKa = 4.42TTTEE374 pKa = 3.97EE375 pKa = 4.28VTPSMLAAWPDD386 pKa = 3.14IYY388 pKa = 11.11SIEE391 pKa = 4.03AQIKK395 pKa = 8.96KK396 pKa = 10.25LADD399 pKa = 3.09RR400 pKa = 11.84VLNRR404 pKa = 11.84KK405 pKa = 7.67YY406 pKa = 10.23QYY408 pKa = 11.41KK409 pKa = 9.89LVEE412 pKa = 4.19RR413 pKa = 11.84DD414 pKa = 3.1FHH416 pKa = 7.5INHH419 pKa = 7.64DD420 pKa = 3.27IQFIVKK426 pKa = 8.81YY427 pKa = 9.13QAA429 pKa = 3.33
MM1 pKa = 6.78NTQLFKK7 pKa = 10.85YY8 pKa = 9.41IKK10 pKa = 9.21KK11 pKa = 9.71FVVKK15 pKa = 10.03VEE17 pKa = 3.64PWTFDD22 pKa = 2.97EE23 pKa = 5.49YY24 pKa = 10.55IASMSSASKK33 pKa = 9.85RR34 pKa = 11.84KK35 pKa = 9.55QYY37 pKa = 11.17SDD39 pKa = 4.97YY40 pKa = 10.82YY41 pKa = 10.97DD42 pKa = 3.07QYY44 pKa = 11.14LRR46 pKa = 11.84NGRR49 pKa = 11.84IPSYY53 pKa = 9.17ITPFTKK59 pKa = 9.42IEE61 pKa = 4.01KK62 pKa = 9.28MSSGKK67 pKa = 10.38YY68 pKa = 8.78KK69 pKa = 10.37APRR72 pKa = 11.84MIQGRR77 pKa = 11.84HH78 pKa = 3.96MVFNLHH84 pKa = 5.66YY85 pKa = 10.28GRR87 pKa = 11.84YY88 pKa = 8.72IKK90 pKa = 10.31PLEE93 pKa = 4.3KK94 pKa = 10.42EE95 pKa = 3.95VTKK98 pKa = 10.79YY99 pKa = 11.08GKK101 pKa = 10.42FSIHH105 pKa = 6.35FGKK108 pKa = 10.89GNYY111 pKa = 7.84NQQAEE116 pKa = 4.8KK117 pKa = 10.42IFKK120 pKa = 10.37LSQKK124 pKa = 9.18YY125 pKa = 9.66KK126 pKa = 10.38YY127 pKa = 9.11YY128 pKa = 10.08TEE130 pKa = 4.94CDD132 pKa = 3.17HH133 pKa = 6.94TSFDD137 pKa = 3.14AHH139 pKa = 5.04VTVEE143 pKa = 4.03QLRR146 pKa = 11.84LTHH149 pKa = 6.96RR150 pKa = 11.84FYY152 pKa = 11.49NSCFPGYY159 pKa = 10.5ASDD162 pKa = 3.78VARR165 pKa = 11.84LAKK168 pKa = 9.5RR169 pKa = 11.84TINNTCISRR178 pKa = 11.84TGDD181 pKa = 2.77IYY183 pKa = 10.73RR184 pKa = 11.84VRR186 pKa = 11.84GSRR189 pKa = 11.84MSGDD193 pKa = 3.15VDD195 pKa = 3.54TGFGNCLINYY205 pKa = 8.01AILKK209 pKa = 8.09AACSNLHH216 pKa = 6.23IKK218 pKa = 10.79CEE220 pKa = 4.17IIVNGDD226 pKa = 3.26DD227 pKa = 4.55SIIFTDD233 pKa = 3.91VPIPTAEE240 pKa = 3.95LSAEE244 pKa = 3.73LRR246 pKa = 11.84KK247 pKa = 10.53YY248 pKa = 11.2NMEE251 pKa = 4.3SKK253 pKa = 10.72VNPSVSDD260 pKa = 2.93IHH262 pKa = 8.17KK263 pKa = 10.75VEE265 pKa = 4.73FCRR268 pKa = 11.84TKK270 pKa = 10.93LVFNASAKK278 pKa = 7.2PTMMIDD284 pKa = 3.68PKK286 pKa = 10.77RR287 pKa = 11.84LIDD290 pKa = 3.89IYY292 pKa = 10.83GMCYY296 pKa = 10.0TISKK300 pKa = 9.92KK301 pKa = 10.43NYY303 pKa = 7.91HH304 pKa = 6.0QYY306 pKa = 11.27LLEE309 pKa = 4.01TAMCNSYY316 pKa = 10.77INCNNPLGVLWAKK329 pKa = 9.96YY330 pKa = 10.11FNIDD334 pKa = 4.39LISYY338 pKa = 8.91KK339 pKa = 10.57KK340 pKa = 10.36NEE342 pKa = 3.77KK343 pKa = 10.54SKK345 pKa = 10.97IKK347 pKa = 10.0ILQNIKK353 pKa = 10.56CLEE356 pKa = 3.84KK357 pKa = 11.06DD358 pKa = 3.93KK359 pKa = 10.64ILKK362 pKa = 9.31MMSLSPDD369 pKa = 3.02EE370 pKa = 4.42TTTEE374 pKa = 3.97EE375 pKa = 4.28VTPSMLAAWPDD386 pKa = 3.14IYY388 pKa = 11.11SIEE391 pKa = 4.03AQIKK395 pKa = 8.96KK396 pKa = 10.25LADD399 pKa = 3.09RR400 pKa = 11.84VLNRR404 pKa = 11.84KK405 pKa = 7.67YY406 pKa = 10.23QYY408 pKa = 11.41KK409 pKa = 9.89LVEE412 pKa = 4.19RR413 pKa = 11.84DD414 pKa = 3.1FHH416 pKa = 7.5INHH419 pKa = 7.64DD420 pKa = 3.27IQFIVKK426 pKa = 8.81YY427 pKa = 9.13QAA429 pKa = 3.33
Molecular weight: 50.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
739 |
310 |
429 |
369.5 |
42.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.954 ± 0.641 | 2.165 ± 0.31 |
5.683 ± 0.431 | 5.413 ± 0.402 |
3.924 ± 0.211 | 4.33 ± 0.466 |
2.706 ± 0.11 | 8.796 ± 0.048 |
10.014 ± 0.37 | 7.307 ± 0.606 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.248 ± 0.012 | 5.683 ± 0.112 |
3.112 ± 0.298 | 3.383 ± 0.093 |
5.142 ± 0.373 | 6.36 ± 0.673 |
5.413 ± 0.04 | 5.007 ± 0.087 |
0.947 ± 0.193 | 5.413 ± 1.409 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
