
Papillomaviridae sp.
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins
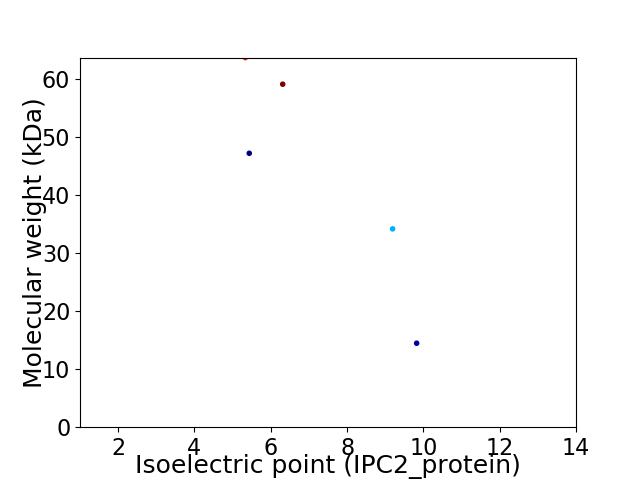
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345N151|A0A345N151_9PAPI Protein E8^E2C OS=Papillomaviridae sp. OX=2052558 PE=3 SV=1
MM1 pKa = 7.48EE2 pKa = 6.46FIDD5 pKa = 4.97TMAEE9 pKa = 4.06CSDD12 pKa = 3.66SGGEE16 pKa = 4.06EE17 pKa = 4.01EE18 pKa = 5.22QEE20 pKa = 4.17VLTVEE25 pKa = 4.48DD26 pKa = 4.72EE27 pKa = 4.46SFIDD31 pKa = 3.29NTQYY35 pKa = 11.1CEE37 pKa = 4.05SPPRR41 pKa = 11.84KK42 pKa = 9.32LLKK45 pKa = 10.0QLCEE49 pKa = 4.14SLEE52 pKa = 4.04KK53 pKa = 10.85SSLEE57 pKa = 3.96KK58 pKa = 10.56KK59 pKa = 8.28YY60 pKa = 11.26AKK62 pKa = 10.0YY63 pKa = 9.95DD64 pKa = 3.47QPGPSVSKK72 pKa = 10.31RR73 pKa = 11.84VLFPEE78 pKa = 4.02RR79 pKa = 11.84HH80 pKa = 5.71VEE82 pKa = 4.1TQEE85 pKa = 3.82DD86 pKa = 4.2SGMLTYY92 pKa = 10.9DD93 pKa = 3.98EE94 pKa = 5.72LSMRR98 pKa = 11.84DD99 pKa = 3.15NYY101 pKa = 10.59EE102 pKa = 3.41INEE105 pKa = 4.17LVNTEE110 pKa = 4.19TLFEE114 pKa = 4.39EE115 pKa = 4.87EE116 pKa = 4.62FIAPVVEE123 pKa = 4.38VQGTLSAQTLEE134 pKa = 4.16QKK136 pKa = 10.65LRR138 pKa = 11.84TSYY141 pKa = 11.13YY142 pKa = 9.36HH143 pKa = 5.75FTAASSPDD151 pKa = 3.58KK152 pKa = 11.03IGVRR156 pKa = 11.84WNDD159 pKa = 2.86VHH161 pKa = 8.97KK162 pKa = 10.87GMKK165 pKa = 9.64SPKK168 pKa = 8.57TMQKK172 pKa = 7.97IWSIFVEE179 pKa = 4.42RR180 pKa = 11.84PVEE183 pKa = 3.98VRR185 pKa = 11.84RR186 pKa = 11.84NHH188 pKa = 6.01PNCGKK193 pKa = 10.13RR194 pKa = 11.84SHH196 pKa = 6.65VPLEE200 pKa = 4.01ALLCNDD206 pKa = 3.66AALLRR211 pKa = 11.84ASSTPHH217 pKa = 5.44HH218 pKa = 6.82VYY220 pKa = 10.89YY221 pKa = 10.83LLEE224 pKa = 4.32YY225 pKa = 9.83TDD227 pKa = 3.71SQRR230 pKa = 11.84SIKK233 pKa = 10.35GLQKK237 pKa = 10.47LLKK240 pKa = 10.26AVGIEE245 pKa = 3.87HH246 pKa = 7.15CLFGVPYY253 pKa = 10.09FKK255 pKa = 10.77QPLTRR260 pKa = 11.84TFLRR264 pKa = 11.84DD265 pKa = 2.96STTPLDD271 pKa = 3.78VKK273 pKa = 10.77RR274 pKa = 11.84DD275 pKa = 3.61VSFLADD281 pKa = 3.76IEE283 pKa = 4.23DD284 pKa = 3.46EE285 pKa = 4.51KK286 pKa = 11.39YY287 pKa = 10.5IKK289 pKa = 10.11QSYY292 pKa = 7.75QFNMEE297 pKa = 4.11EE298 pKa = 4.21LISYY302 pKa = 9.84CEE304 pKa = 3.94SSEE307 pKa = 4.24PEE309 pKa = 4.08TVQQLIYY316 pKa = 10.39RR317 pKa = 11.84YY318 pKa = 7.93TSEE321 pKa = 4.97AKK323 pKa = 10.48HH324 pKa = 6.12GDD326 pKa = 3.41PNAIAWRR333 pKa = 11.84NTTSCMNHH341 pKa = 6.13AKK343 pKa = 10.29NAFSMWKK350 pKa = 8.26STVQGEE356 pKa = 4.27MLDD359 pKa = 4.4LSLSDD364 pKa = 4.6YY365 pKa = 10.18ILHH368 pKa = 7.07RR369 pKa = 11.84ISEE372 pKa = 4.37NEE374 pKa = 3.81GGDD377 pKa = 3.38AAKK380 pKa = 10.3VRR382 pKa = 11.84RR383 pKa = 11.84LFTIQGILEE392 pKa = 3.99IDD394 pKa = 3.96FLNTLRR400 pKa = 11.84KK401 pKa = 8.98WLVGQHH407 pKa = 6.03KK408 pKa = 10.27YY409 pKa = 10.76NVIVLVGPGNTGKK422 pKa = 10.64SLFTEE427 pKa = 4.51ALMKK431 pKa = 10.74CLGGAFLSWHH441 pKa = 7.25KK442 pKa = 11.01DD443 pKa = 3.12NQYY446 pKa = 9.46WKK448 pKa = 10.85SPILGSRR455 pKa = 11.84FCCIDD460 pKa = 4.94DD461 pKa = 3.45ITLAGWKK468 pKa = 10.12NLDD471 pKa = 3.33EE472 pKa = 4.62CEE474 pKa = 4.26RR475 pKa = 11.84RR476 pKa = 11.84ALDD479 pKa = 3.79GGIVTINKK487 pKa = 9.43KK488 pKa = 8.32FTQPTEE494 pKa = 4.18VKK496 pKa = 10.39FPPCLITTNTILTDD510 pKa = 3.46ADD512 pKa = 4.29FSFLKK517 pKa = 10.6NRR519 pKa = 11.84LVWYY523 pKa = 8.33TFDD526 pKa = 3.44KK527 pKa = 11.3VLVTRR532 pKa = 11.84DD533 pKa = 3.52GLAEE537 pKa = 4.13AAVSTADD544 pKa = 3.37CAAWLLLNRR553 pKa = 11.84NTLDD557 pKa = 3.33LL558 pKa = 4.63
MM1 pKa = 7.48EE2 pKa = 6.46FIDD5 pKa = 4.97TMAEE9 pKa = 4.06CSDD12 pKa = 3.66SGGEE16 pKa = 4.06EE17 pKa = 4.01EE18 pKa = 5.22QEE20 pKa = 4.17VLTVEE25 pKa = 4.48DD26 pKa = 4.72EE27 pKa = 4.46SFIDD31 pKa = 3.29NTQYY35 pKa = 11.1CEE37 pKa = 4.05SPPRR41 pKa = 11.84KK42 pKa = 9.32LLKK45 pKa = 10.0QLCEE49 pKa = 4.14SLEE52 pKa = 4.04KK53 pKa = 10.85SSLEE57 pKa = 3.96KK58 pKa = 10.56KK59 pKa = 8.28YY60 pKa = 11.26AKK62 pKa = 10.0YY63 pKa = 9.95DD64 pKa = 3.47QPGPSVSKK72 pKa = 10.31RR73 pKa = 11.84VLFPEE78 pKa = 4.02RR79 pKa = 11.84HH80 pKa = 5.71VEE82 pKa = 4.1TQEE85 pKa = 3.82DD86 pKa = 4.2SGMLTYY92 pKa = 10.9DD93 pKa = 3.98EE94 pKa = 5.72LSMRR98 pKa = 11.84DD99 pKa = 3.15NYY101 pKa = 10.59EE102 pKa = 3.41INEE105 pKa = 4.17LVNTEE110 pKa = 4.19TLFEE114 pKa = 4.39EE115 pKa = 4.87EE116 pKa = 4.62FIAPVVEE123 pKa = 4.38VQGTLSAQTLEE134 pKa = 4.16QKK136 pKa = 10.65LRR138 pKa = 11.84TSYY141 pKa = 11.13YY142 pKa = 9.36HH143 pKa = 5.75FTAASSPDD151 pKa = 3.58KK152 pKa = 11.03IGVRR156 pKa = 11.84WNDD159 pKa = 2.86VHH161 pKa = 8.97KK162 pKa = 10.87GMKK165 pKa = 9.64SPKK168 pKa = 8.57TMQKK172 pKa = 7.97IWSIFVEE179 pKa = 4.42RR180 pKa = 11.84PVEE183 pKa = 3.98VRR185 pKa = 11.84RR186 pKa = 11.84NHH188 pKa = 6.01PNCGKK193 pKa = 10.13RR194 pKa = 11.84SHH196 pKa = 6.65VPLEE200 pKa = 4.01ALLCNDD206 pKa = 3.66AALLRR211 pKa = 11.84ASSTPHH217 pKa = 5.44HH218 pKa = 6.82VYY220 pKa = 10.89YY221 pKa = 10.83LLEE224 pKa = 4.32YY225 pKa = 9.83TDD227 pKa = 3.71SQRR230 pKa = 11.84SIKK233 pKa = 10.35GLQKK237 pKa = 10.47LLKK240 pKa = 10.26AVGIEE245 pKa = 3.87HH246 pKa = 7.15CLFGVPYY253 pKa = 10.09FKK255 pKa = 10.77QPLTRR260 pKa = 11.84TFLRR264 pKa = 11.84DD265 pKa = 2.96STTPLDD271 pKa = 3.78VKK273 pKa = 10.77RR274 pKa = 11.84DD275 pKa = 3.61VSFLADD281 pKa = 3.76IEE283 pKa = 4.23DD284 pKa = 3.46EE285 pKa = 4.51KK286 pKa = 11.39YY287 pKa = 10.5IKK289 pKa = 10.11QSYY292 pKa = 7.75QFNMEE297 pKa = 4.11EE298 pKa = 4.21LISYY302 pKa = 9.84CEE304 pKa = 3.94SSEE307 pKa = 4.24PEE309 pKa = 4.08TVQQLIYY316 pKa = 10.39RR317 pKa = 11.84YY318 pKa = 7.93TSEE321 pKa = 4.97AKK323 pKa = 10.48HH324 pKa = 6.12GDD326 pKa = 3.41PNAIAWRR333 pKa = 11.84NTTSCMNHH341 pKa = 6.13AKK343 pKa = 10.29NAFSMWKK350 pKa = 8.26STVQGEE356 pKa = 4.27MLDD359 pKa = 4.4LSLSDD364 pKa = 4.6YY365 pKa = 10.18ILHH368 pKa = 7.07RR369 pKa = 11.84ISEE372 pKa = 4.37NEE374 pKa = 3.81GGDD377 pKa = 3.38AAKK380 pKa = 10.3VRR382 pKa = 11.84RR383 pKa = 11.84LFTIQGILEE392 pKa = 3.99IDD394 pKa = 3.96FLNTLRR400 pKa = 11.84KK401 pKa = 8.98WLVGQHH407 pKa = 6.03KK408 pKa = 10.27YY409 pKa = 10.76NVIVLVGPGNTGKK422 pKa = 10.64SLFTEE427 pKa = 4.51ALMKK431 pKa = 10.74CLGGAFLSWHH441 pKa = 7.25KK442 pKa = 11.01DD443 pKa = 3.12NQYY446 pKa = 9.46WKK448 pKa = 10.85SPILGSRR455 pKa = 11.84FCCIDD460 pKa = 4.94DD461 pKa = 3.45ITLAGWKK468 pKa = 10.12NLDD471 pKa = 3.33EE472 pKa = 4.62CEE474 pKa = 4.26RR475 pKa = 11.84RR476 pKa = 11.84ALDD479 pKa = 3.79GGIVTINKK487 pKa = 9.43KK488 pKa = 8.32FTQPTEE494 pKa = 4.18VKK496 pKa = 10.39FPPCLITTNTILTDD510 pKa = 3.46ADD512 pKa = 4.29FSFLKK517 pKa = 10.6NRR519 pKa = 11.84LVWYY523 pKa = 8.33TFDD526 pKa = 3.44KK527 pKa = 11.3VLVTRR532 pKa = 11.84DD533 pKa = 3.52GLAEE537 pKa = 4.13AAVSTADD544 pKa = 3.37CAAWLLLNRR553 pKa = 11.84NTLDD557 pKa = 3.33LL558 pKa = 4.63
Molecular weight: 63.76 kDa
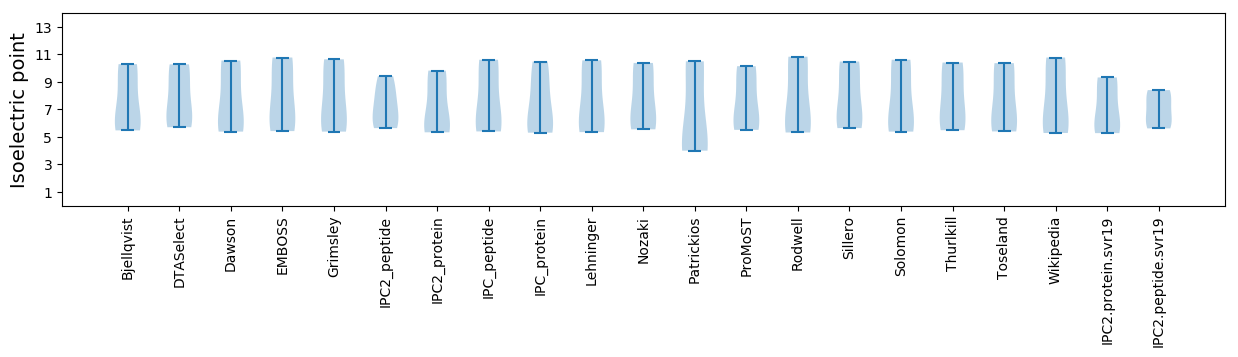
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345N148|A0A345N148_9PAPI L2 protein OS=Papillomaviridae sp. OX=2052558 PE=4 SV=1
MM1 pKa = 7.4VNGKK5 pKa = 7.98QEE7 pKa = 3.81QMDD10 pKa = 3.55IGVLLICLMDD20 pKa = 4.4LFQHH24 pKa = 6.02SLRR27 pKa = 11.84LKK29 pKa = 10.35QVIQMQPLISITDD42 pKa = 3.63SLILSNRR49 pKa = 11.84GLVLHH54 pKa = 6.55HH55 pKa = 6.31HH56 pKa = 5.46QKK58 pKa = 10.53SRR60 pKa = 11.84LEE62 pKa = 4.12VIIIFHH68 pKa = 6.04LTLYY72 pKa = 10.4IILIRR77 pKa = 11.84NMYY80 pKa = 8.11ITRR83 pKa = 11.84KK84 pKa = 8.79YY85 pKa = 9.81RR86 pKa = 11.84RR87 pKa = 11.84NVKK90 pKa = 9.11NAKK93 pKa = 9.21ARR95 pKa = 11.84NVLSMMCNYY104 pKa = 9.98SVSRR108 pKa = 11.84TYY110 pKa = 10.72SSGVRR115 pKa = 11.84VHH117 pKa = 7.03RR118 pKa = 11.84GWCTYY123 pKa = 10.46
MM1 pKa = 7.4VNGKK5 pKa = 7.98QEE7 pKa = 3.81QMDD10 pKa = 3.55IGVLLICLMDD20 pKa = 4.4LFQHH24 pKa = 6.02SLRR27 pKa = 11.84LKK29 pKa = 10.35QVIQMQPLISITDD42 pKa = 3.63SLILSNRR49 pKa = 11.84GLVLHH54 pKa = 6.55HH55 pKa = 6.31HH56 pKa = 5.46QKK58 pKa = 10.53SRR60 pKa = 11.84LEE62 pKa = 4.12VIIIFHH68 pKa = 6.04LTLYY72 pKa = 10.4IILIRR77 pKa = 11.84NMYY80 pKa = 8.11ITRR83 pKa = 11.84KK84 pKa = 8.79YY85 pKa = 9.81RR86 pKa = 11.84RR87 pKa = 11.84NVKK90 pKa = 9.11NAKK93 pKa = 9.21ARR95 pKa = 11.84NVLSMMCNYY104 pKa = 9.98SVSRR108 pKa = 11.84TYY110 pKa = 10.72SSGVRR115 pKa = 11.84VHH117 pKa = 7.03RR118 pKa = 11.84GWCTYY123 pKa = 10.46
Molecular weight: 14.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1936 |
123 |
558 |
387.2 |
43.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.888 ± 0.574 | 1.498 ± 0.418 |
6.043 ± 0.689 | 6.508 ± 0.602 |
3.874 ± 0.233 | 6.198 ± 0.53 |
3.099 ± 0.458 | 5.63 ± 0.506 |
7.128 ± 1.11 | 8.523 ± 1.133 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.963 ± 0.502 | 4.029 ± 0.353 |
6.612 ± 1.7 | 3.306 ± 0.497 |
5.837 ± 0.74 | 6.56 ± 0.575 |
6.508 ± 0.246 | 6.043 ± 0.294 |
1.343 ± 0.177 | 3.409 ± 0.299 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
