
Synechococcus phage ACG-2014i
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 5.6
Get precalculated fractions of proteins
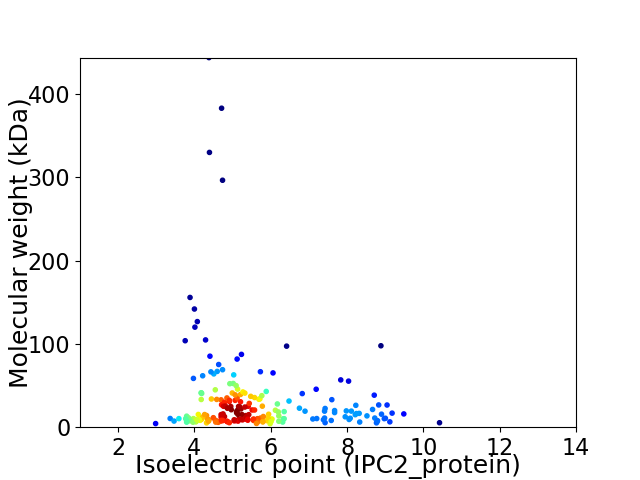
Virtual 2D-PAGE plot for 212 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E3FGE6|A0A0E3FGE6_9CAUD Peptidase_S8 domain-containing protein OS=Synechococcus phage ACG-2014i OX=1493513 GN=Syn7803US120_170 PE=4 SV=1
MM1 pKa = 7.43SKK3 pKa = 10.01ILANQIANYY12 pKa = 9.63GDD14 pKa = 3.53NSPVEE19 pKa = 4.3VKK21 pKa = 10.66EE22 pKa = 4.11GVNIPAGKK30 pKa = 8.71PLQAAGVAGTSGQVLTATGASIQWTTPFDD59 pKa = 3.68GSYY62 pKa = 9.46LTLTNKK68 pKa = 8.43PTIPAAQINADD79 pKa = 3.45WNASGGSVAAILNKK93 pKa = 9.8PVIPAQPSIVLTAAGSSTLTYY114 pKa = 10.25NQANGEE120 pKa = 4.31FTFTPPDD127 pKa = 3.67FSSYY131 pKa = 9.15LTTYY135 pKa = 10.41TEE137 pKa = 3.91TDD139 pKa = 4.26PIFLASPAYY148 pKa = 10.29QITNQNRR155 pKa = 11.84TDD157 pKa = 3.07WGLAYY162 pKa = 10.44SWGDD166 pKa = 3.3HH167 pKa = 5.43SQAGYY172 pKa = 7.87ITTYY176 pKa = 10.68TEE178 pKa = 3.7TDD180 pKa = 3.48PVFTASVAAGITTQNKK196 pKa = 7.69TNWDD200 pKa = 3.34TSYY203 pKa = 11.26GWGNHH208 pKa = 5.07AAQGYY213 pKa = 5.2ITSYY217 pKa = 11.24SEE219 pKa = 3.72TDD221 pKa = 3.46PVFTASVAAGITLTNVSNWDD241 pKa = 3.33TAYY244 pKa = 9.24TWGDD248 pKa = 3.23HH249 pKa = 5.45GQAGYY254 pKa = 8.44LTDD257 pKa = 4.68LLSSSIEE264 pKa = 3.97LLSDD268 pKa = 3.44VAISGPVADD277 pKa = 5.75QLLKK281 pKa = 11.2YY282 pKa = 10.55NGSNWINYY290 pKa = 6.46TPSYY294 pKa = 9.98LEE296 pKa = 4.99SYY298 pKa = 10.29TEE300 pKa = 4.04TDD302 pKa = 3.08TLASITARR310 pKa = 11.84GASTTTTVTLTDD322 pKa = 4.38LNVSGNLNVLGTTTTNNVSTLNVTNNEE349 pKa = 3.42IVLNEE354 pKa = 3.98NQASGGLDD362 pKa = 2.79ALLTNEE368 pKa = 4.93RR369 pKa = 11.84GTDD372 pKa = 3.19ADD374 pKa = 5.36VSIKK378 pKa = 10.13WNEE381 pKa = 4.01TTDD384 pKa = 2.62RR385 pKa = 11.84WQFTNDD391 pKa = 2.19GSTYY395 pKa = 10.5YY396 pKa = 10.61NVAINASEE404 pKa = 4.09LTNDD408 pKa = 3.47AGYY411 pKa = 8.41LTTSSSINSLGDD423 pKa = 3.38VTISTPTSGDD433 pKa = 3.36VLSYY437 pKa = 11.13NGSDD441 pKa = 3.13WVNSPASITAKK452 pKa = 10.73ASISDD457 pKa = 4.12AAPSSPSPGDD467 pKa = 3.32MWWKK471 pKa = 10.25SDD473 pKa = 3.56EE474 pKa = 4.07GTMKK478 pKa = 10.13VWYY481 pKa = 9.67DD482 pKa = 4.39DD483 pKa = 4.18GNTAQWVDD491 pKa = 3.54ASPVGDD497 pKa = 3.59PFEE500 pKa = 4.24NVYY503 pKa = 11.15ASVAFFPGANVNTGAFAYY521 pKa = 10.44SEE523 pKa = 4.42ATGAMYY529 pKa = 10.71YY530 pKa = 10.4SNSVSWTSQRR540 pKa = 11.84LVTTNSSTSSDD551 pKa = 3.62FATLLANTQLTYY563 pKa = 10.73SVSAIDD569 pKa = 3.73YY570 pKa = 6.99TTGGTAEE577 pKa = 3.9YY578 pKa = 10.16NAARR582 pKa = 11.84KK583 pKa = 8.93IVRR586 pKa = 11.84LSDD589 pKa = 3.6SQGVTGDD596 pKa = 3.93IILTAGTGLSIVKK609 pKa = 9.98SNNEE613 pKa = 3.28ITFNNDD619 pKa = 2.5VVDD622 pKa = 3.82TTYY625 pKa = 11.04GISVEE630 pKa = 4.09ASSGANSILRR640 pKa = 11.84LTDD643 pKa = 3.29SQGVLDD649 pKa = 4.49DD650 pKa = 4.12VVFAGADD657 pKa = 3.51GLVVEE662 pKa = 4.87NTDD665 pKa = 3.22ANTFTFRR672 pKa = 11.84APDD675 pKa = 3.31ISSQFYY681 pKa = 10.15TDD683 pKa = 4.82EE684 pKa = 4.17EE685 pKa = 4.35AQDD688 pKa = 3.96AVATMFANGTHH699 pKa = 5.55TNITFTYY706 pKa = 10.57DD707 pKa = 3.17DD708 pKa = 4.25TNNSLSATAQAGGGGGGGTTYY729 pKa = 11.39DD730 pKa = 3.51LVGSNTNSNNAIITLLDD747 pKa = 3.77ANNNEE752 pKa = 4.49DD753 pKa = 3.99KK754 pKa = 11.16IEE756 pKa = 4.01IAGGGGTDD764 pKa = 4.13VSWDD768 pKa = 3.67GPNEE772 pKa = 4.41RR773 pKa = 11.84ITVSSTAPVQSDD785 pKa = 2.55WDD787 pKa = 3.82ATTGLAQILNKK798 pKa = 10.04PSIPSAYY805 pKa = 10.13ALPTAAAGTLGGIKK819 pKa = 10.14VGANLSIDD827 pKa = 3.41GDD829 pKa = 4.59GILSANAGGYY839 pKa = 6.9TLPAADD845 pKa = 3.84ATSLGGIKK853 pKa = 10.15VGSGLSIDD861 pKa = 3.98GNGVLTATGGSSVPSIQDD879 pKa = 3.24LSGTTASLAADD890 pKa = 3.54QSAEE894 pKa = 4.1LNITGYY900 pKa = 10.39KK901 pKa = 9.96AYY903 pKa = 10.93SLFKK907 pKa = 10.12VTTDD911 pKa = 2.8AEE913 pKa = 4.11AWVRR917 pKa = 11.84VYY919 pKa = 11.33VDD921 pKa = 3.58DD922 pKa = 5.58ASRR925 pKa = 11.84DD926 pKa = 3.42ADD928 pKa = 3.55NTRR931 pKa = 11.84SEE933 pKa = 5.02GEE935 pKa = 4.23DD936 pKa = 3.44PTPGSGVISEE946 pKa = 4.3VRR948 pKa = 11.84TSGAEE953 pKa = 3.87SVLISPGIMGFNNDD967 pKa = 3.48SPRR970 pKa = 11.84TDD972 pKa = 3.49TIYY975 pKa = 11.08LSVTNRR981 pKa = 11.84SGSATTITVTLTALQIGEE999 pKa = 4.14
MM1 pKa = 7.43SKK3 pKa = 10.01ILANQIANYY12 pKa = 9.63GDD14 pKa = 3.53NSPVEE19 pKa = 4.3VKK21 pKa = 10.66EE22 pKa = 4.11GVNIPAGKK30 pKa = 8.71PLQAAGVAGTSGQVLTATGASIQWTTPFDD59 pKa = 3.68GSYY62 pKa = 9.46LTLTNKK68 pKa = 8.43PTIPAAQINADD79 pKa = 3.45WNASGGSVAAILNKK93 pKa = 9.8PVIPAQPSIVLTAAGSSTLTYY114 pKa = 10.25NQANGEE120 pKa = 4.31FTFTPPDD127 pKa = 3.67FSSYY131 pKa = 9.15LTTYY135 pKa = 10.41TEE137 pKa = 3.91TDD139 pKa = 4.26PIFLASPAYY148 pKa = 10.29QITNQNRR155 pKa = 11.84TDD157 pKa = 3.07WGLAYY162 pKa = 10.44SWGDD166 pKa = 3.3HH167 pKa = 5.43SQAGYY172 pKa = 7.87ITTYY176 pKa = 10.68TEE178 pKa = 3.7TDD180 pKa = 3.48PVFTASVAAGITTQNKK196 pKa = 7.69TNWDD200 pKa = 3.34TSYY203 pKa = 11.26GWGNHH208 pKa = 5.07AAQGYY213 pKa = 5.2ITSYY217 pKa = 11.24SEE219 pKa = 3.72TDD221 pKa = 3.46PVFTASVAAGITLTNVSNWDD241 pKa = 3.33TAYY244 pKa = 9.24TWGDD248 pKa = 3.23HH249 pKa = 5.45GQAGYY254 pKa = 8.44LTDD257 pKa = 4.68LLSSSIEE264 pKa = 3.97LLSDD268 pKa = 3.44VAISGPVADD277 pKa = 5.75QLLKK281 pKa = 11.2YY282 pKa = 10.55NGSNWINYY290 pKa = 6.46TPSYY294 pKa = 9.98LEE296 pKa = 4.99SYY298 pKa = 10.29TEE300 pKa = 4.04TDD302 pKa = 3.08TLASITARR310 pKa = 11.84GASTTTTVTLTDD322 pKa = 4.38LNVSGNLNVLGTTTTNNVSTLNVTNNEE349 pKa = 3.42IVLNEE354 pKa = 3.98NQASGGLDD362 pKa = 2.79ALLTNEE368 pKa = 4.93RR369 pKa = 11.84GTDD372 pKa = 3.19ADD374 pKa = 5.36VSIKK378 pKa = 10.13WNEE381 pKa = 4.01TTDD384 pKa = 2.62RR385 pKa = 11.84WQFTNDD391 pKa = 2.19GSTYY395 pKa = 10.5YY396 pKa = 10.61NVAINASEE404 pKa = 4.09LTNDD408 pKa = 3.47AGYY411 pKa = 8.41LTTSSSINSLGDD423 pKa = 3.38VTISTPTSGDD433 pKa = 3.36VLSYY437 pKa = 11.13NGSDD441 pKa = 3.13WVNSPASITAKK452 pKa = 10.73ASISDD457 pKa = 4.12AAPSSPSPGDD467 pKa = 3.32MWWKK471 pKa = 10.25SDD473 pKa = 3.56EE474 pKa = 4.07GTMKK478 pKa = 10.13VWYY481 pKa = 9.67DD482 pKa = 4.39DD483 pKa = 4.18GNTAQWVDD491 pKa = 3.54ASPVGDD497 pKa = 3.59PFEE500 pKa = 4.24NVYY503 pKa = 11.15ASVAFFPGANVNTGAFAYY521 pKa = 10.44SEE523 pKa = 4.42ATGAMYY529 pKa = 10.71YY530 pKa = 10.4SNSVSWTSQRR540 pKa = 11.84LVTTNSSTSSDD551 pKa = 3.62FATLLANTQLTYY563 pKa = 10.73SVSAIDD569 pKa = 3.73YY570 pKa = 6.99TTGGTAEE577 pKa = 3.9YY578 pKa = 10.16NAARR582 pKa = 11.84KK583 pKa = 8.93IVRR586 pKa = 11.84LSDD589 pKa = 3.6SQGVTGDD596 pKa = 3.93IILTAGTGLSIVKK609 pKa = 9.98SNNEE613 pKa = 3.28ITFNNDD619 pKa = 2.5VVDD622 pKa = 3.82TTYY625 pKa = 11.04GISVEE630 pKa = 4.09ASSGANSILRR640 pKa = 11.84LTDD643 pKa = 3.29SQGVLDD649 pKa = 4.49DD650 pKa = 4.12VVFAGADD657 pKa = 3.51GLVVEE662 pKa = 4.87NTDD665 pKa = 3.22ANTFTFRR672 pKa = 11.84APDD675 pKa = 3.31ISSQFYY681 pKa = 10.15TDD683 pKa = 4.82EE684 pKa = 4.17EE685 pKa = 4.35AQDD688 pKa = 3.96AVATMFANGTHH699 pKa = 5.55TNITFTYY706 pKa = 10.57DD707 pKa = 3.17DD708 pKa = 4.25TNNSLSATAQAGGGGGGGTTYY729 pKa = 11.39DD730 pKa = 3.51LVGSNTNSNNAIITLLDD747 pKa = 3.77ANNNEE752 pKa = 4.49DD753 pKa = 3.99KK754 pKa = 11.16IEE756 pKa = 4.01IAGGGGTDD764 pKa = 4.13VSWDD768 pKa = 3.67GPNEE772 pKa = 4.41RR773 pKa = 11.84ITVSSTAPVQSDD785 pKa = 2.55WDD787 pKa = 3.82ATTGLAQILNKK798 pKa = 10.04PSIPSAYY805 pKa = 10.13ALPTAAAGTLGGIKK819 pKa = 10.14VGANLSIDD827 pKa = 3.41GDD829 pKa = 4.59GILSANAGGYY839 pKa = 6.9TLPAADD845 pKa = 3.84ATSLGGIKK853 pKa = 10.15VGSGLSIDD861 pKa = 3.98GNGVLTATGGSSVPSIQDD879 pKa = 3.24LSGTTASLAADD890 pKa = 3.54QSAEE894 pKa = 4.1LNITGYY900 pKa = 10.39KK901 pKa = 9.96AYY903 pKa = 10.93SLFKK907 pKa = 10.12VTTDD911 pKa = 2.8AEE913 pKa = 4.11AWVRR917 pKa = 11.84VYY919 pKa = 11.33VDD921 pKa = 3.58DD922 pKa = 5.58ASRR925 pKa = 11.84DD926 pKa = 3.42ADD928 pKa = 3.55NTRR931 pKa = 11.84SEE933 pKa = 5.02GEE935 pKa = 4.23DD936 pKa = 3.44PTPGSGVISEE946 pKa = 4.3VRR948 pKa = 11.84TSGAEE953 pKa = 3.87SVLISPGIMGFNNDD967 pKa = 3.48SPRR970 pKa = 11.84TDD972 pKa = 3.49TIYY975 pKa = 11.08LSVTNRR981 pKa = 11.84SGSATTITVTLTALQIGEE999 pKa = 4.14
Molecular weight: 103.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3FGA2|A0A0E3FGA2_9CAUD Plastocyanine OS=Synechococcus phage ACG-2014i OX=1493513 GN=Syn7803US120_95 PE=3 SV=1
MM1 pKa = 7.39AKK3 pKa = 10.09SRR5 pKa = 11.84VGLSGAEE12 pKa = 4.21TIEE15 pKa = 4.65SIPKK19 pKa = 8.64RR20 pKa = 11.84TRR22 pKa = 11.84QGRR25 pKa = 11.84GKK27 pKa = 7.68HH28 pKa = 4.65TKK30 pKa = 8.57YY31 pKa = 9.26TATSRR36 pKa = 11.84NGAKK40 pKa = 9.61KK41 pKa = 10.28RR42 pKa = 11.84YY43 pKa = 8.62RR44 pKa = 11.84GQGKK48 pKa = 9.26GG49 pKa = 3.22
MM1 pKa = 7.39AKK3 pKa = 10.09SRR5 pKa = 11.84VGLSGAEE12 pKa = 4.21TIEE15 pKa = 4.65SIPKK19 pKa = 8.64RR20 pKa = 11.84TRR22 pKa = 11.84QGRR25 pKa = 11.84GKK27 pKa = 7.68HH28 pKa = 4.65TKK30 pKa = 8.57YY31 pKa = 9.26TATSRR36 pKa = 11.84NGAKK40 pKa = 9.61KK41 pKa = 10.28RR42 pKa = 11.84YY43 pKa = 8.62RR44 pKa = 11.84GQGKK48 pKa = 9.26GG49 pKa = 3.22
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
60288 |
38 |
4102 |
284.4 |
31.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.738 ± 0.257 | 0.841 ± 0.082 |
6.688 ± 0.121 | 5.787 ± 0.278 |
4.296 ± 0.113 | 8.265 ± 0.381 |
1.358 ± 0.124 | 6.515 ± 0.18 |
5.319 ± 0.343 | 7.083 ± 0.174 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.939 ± 0.201 | 6.17 ± 0.19 |
3.956 ± 0.125 | 3.531 ± 0.101 |
3.812 ± 0.119 | 7.681 ± 0.274 |
7.862 ± 0.32 | 6.726 ± 0.176 |
1.148 ± 0.083 | 4.279 ± 0.153 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
