
Microbacterium pygmaeum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
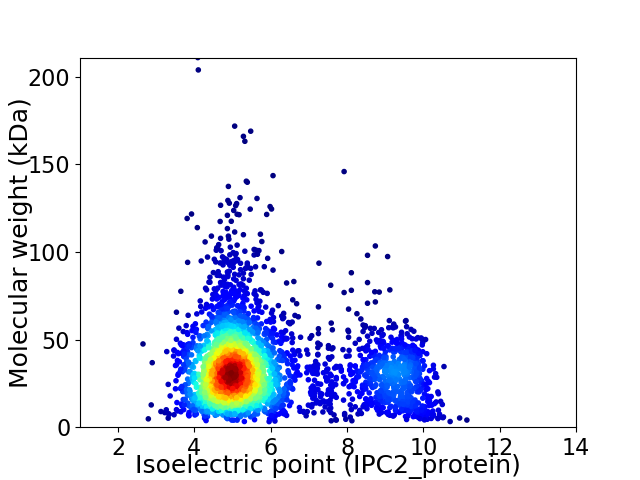
Virtual 2D-PAGE plot for 3412 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G7WS51|A0A1G7WS51_9MICO Putative adhesin OS=Microbacterium pygmaeum OX=370764 GN=SAMN04489810_1180 PE=4 SV=1
MM1 pKa = 7.34IRR3 pKa = 11.84WKK5 pKa = 10.39KK6 pKa = 9.6AAAAAAIAVTAALALSSCAGGATDD30 pKa = 4.8GGGEE34 pKa = 4.23GGSGGTLTLGAIAAPTTFDD53 pKa = 3.75PAGSEE58 pKa = 3.78WGNRR62 pKa = 11.84SPFYY66 pKa = 10.45QAVFDD71 pKa = 4.24TLLLATPEE79 pKa = 4.18GTIEE83 pKa = 3.82PWLATEE89 pKa = 4.74WSYY92 pKa = 12.27NDD94 pKa = 4.13DD95 pKa = 3.76NTVLTLTIRR104 pKa = 11.84DD105 pKa = 4.07DD106 pKa = 3.62VTFTDD111 pKa = 5.55GSALTADD118 pKa = 3.61VVVGNLQRR126 pKa = 11.84FKK128 pKa = 11.28DD129 pKa = 3.91GTSPDD134 pKa = 3.15AGYY137 pKa = 9.51FAGVASFEE145 pKa = 4.52APDD148 pKa = 3.76DD149 pKa = 3.88TTVVITLSAPDD160 pKa = 4.25PAMLDD165 pKa = 3.53YY166 pKa = 10.14LTRR169 pKa = 11.84DD170 pKa = 3.42PGLVGAEE177 pKa = 4.07ANFDD181 pKa = 3.85NPDD184 pKa = 3.44AATTPIGSGPYY195 pKa = 10.23VLDD198 pKa = 3.31TAATVTGTTYY208 pKa = 11.02AYY210 pKa = 8.09TKK212 pKa = 10.91NPDD215 pKa = 2.94YY216 pKa = 10.7WNPDD220 pKa = 2.95VQHH223 pKa = 6.58YY224 pKa = 10.47DD225 pKa = 3.17NLVINTLTDD234 pKa = 3.17PTAALNAIKK243 pKa = 10.33AGEE246 pKa = 4.02ANGVKK251 pKa = 10.06LANNDD256 pKa = 3.51ALDD259 pKa = 3.98EE260 pKa = 4.35VEE262 pKa = 4.29GAGWTVNANEE272 pKa = 5.21LDD274 pKa = 3.93FQGLLLLDD282 pKa = 4.02RR283 pKa = 11.84AGTMDD288 pKa = 4.61PALADD293 pKa = 3.49VKK295 pKa = 10.74VRR297 pKa = 11.84QAINYY302 pKa = 8.64AFDD305 pKa = 4.46RR306 pKa = 11.84EE307 pKa = 4.29GLLQATQFGNGTVTTQVFPATSDD330 pKa = 3.41AYY332 pKa = 10.76DD333 pKa = 4.06PEE335 pKa = 5.38LDD337 pKa = 3.44EE338 pKa = 5.23YY339 pKa = 10.33YY340 pKa = 10.49TYY342 pKa = 11.2DD343 pKa = 3.5PEE345 pKa = 4.43KK346 pKa = 10.79AKK348 pKa = 10.97SLLAEE353 pKa = 4.42AGYY356 pKa = 11.36ADD358 pKa = 4.69GLTISMPSVSVLGATTYY375 pKa = 10.89TLVAQQLADD384 pKa = 3.05IGITVEE390 pKa = 4.19QVDD393 pKa = 4.05VPMGNFIADD402 pKa = 4.6LLAPKK407 pKa = 10.34YY408 pKa = 8.63PASFMALEE416 pKa = 4.41QNPDD420 pKa = 2.7WQLIQFMIAPTAVFNPFKK438 pKa = 11.04YY439 pKa = 10.22SDD441 pKa = 3.94PQVDD445 pKa = 4.0EE446 pKa = 4.83YY447 pKa = 10.85IQEE450 pKa = 3.82IQYY453 pKa = 11.24GDD455 pKa = 4.11EE456 pKa = 3.94ATQASVAKK464 pKa = 9.68EE465 pKa = 3.68LNTYY469 pKa = 9.78IVEE472 pKa = 4.41QGWFAPFFRR481 pKa = 11.84VQGSVATDD489 pKa = 3.11ANTTVEE495 pKa = 4.19MLPTNAYY502 pKa = 8.08PAIYY506 pKa = 10.15DD507 pKa = 3.79FQPKK511 pKa = 7.63QQ512 pKa = 3.15
MM1 pKa = 7.34IRR3 pKa = 11.84WKK5 pKa = 10.39KK6 pKa = 9.6AAAAAAIAVTAALALSSCAGGATDD30 pKa = 4.8GGGEE34 pKa = 4.23GGSGGTLTLGAIAAPTTFDD53 pKa = 3.75PAGSEE58 pKa = 3.78WGNRR62 pKa = 11.84SPFYY66 pKa = 10.45QAVFDD71 pKa = 4.24TLLLATPEE79 pKa = 4.18GTIEE83 pKa = 3.82PWLATEE89 pKa = 4.74WSYY92 pKa = 12.27NDD94 pKa = 4.13DD95 pKa = 3.76NTVLTLTIRR104 pKa = 11.84DD105 pKa = 4.07DD106 pKa = 3.62VTFTDD111 pKa = 5.55GSALTADD118 pKa = 3.61VVVGNLQRR126 pKa = 11.84FKK128 pKa = 11.28DD129 pKa = 3.91GTSPDD134 pKa = 3.15AGYY137 pKa = 9.51FAGVASFEE145 pKa = 4.52APDD148 pKa = 3.76DD149 pKa = 3.88TTVVITLSAPDD160 pKa = 4.25PAMLDD165 pKa = 3.53YY166 pKa = 10.14LTRR169 pKa = 11.84DD170 pKa = 3.42PGLVGAEE177 pKa = 4.07ANFDD181 pKa = 3.85NPDD184 pKa = 3.44AATTPIGSGPYY195 pKa = 10.23VLDD198 pKa = 3.31TAATVTGTTYY208 pKa = 11.02AYY210 pKa = 8.09TKK212 pKa = 10.91NPDD215 pKa = 2.94YY216 pKa = 10.7WNPDD220 pKa = 2.95VQHH223 pKa = 6.58YY224 pKa = 10.47DD225 pKa = 3.17NLVINTLTDD234 pKa = 3.17PTAALNAIKK243 pKa = 10.33AGEE246 pKa = 4.02ANGVKK251 pKa = 10.06LANNDD256 pKa = 3.51ALDD259 pKa = 3.98EE260 pKa = 4.35VEE262 pKa = 4.29GAGWTVNANEE272 pKa = 5.21LDD274 pKa = 3.93FQGLLLLDD282 pKa = 4.02RR283 pKa = 11.84AGTMDD288 pKa = 4.61PALADD293 pKa = 3.49VKK295 pKa = 10.74VRR297 pKa = 11.84QAINYY302 pKa = 8.64AFDD305 pKa = 4.46RR306 pKa = 11.84EE307 pKa = 4.29GLLQATQFGNGTVTTQVFPATSDD330 pKa = 3.41AYY332 pKa = 10.76DD333 pKa = 4.06PEE335 pKa = 5.38LDD337 pKa = 3.44EE338 pKa = 5.23YY339 pKa = 10.33YY340 pKa = 10.49TYY342 pKa = 11.2DD343 pKa = 3.5PEE345 pKa = 4.43KK346 pKa = 10.79AKK348 pKa = 10.97SLLAEE353 pKa = 4.42AGYY356 pKa = 11.36ADD358 pKa = 4.69GLTISMPSVSVLGATTYY375 pKa = 10.89TLVAQQLADD384 pKa = 3.05IGITVEE390 pKa = 4.19QVDD393 pKa = 4.05VPMGNFIADD402 pKa = 4.6LLAPKK407 pKa = 10.34YY408 pKa = 8.63PASFMALEE416 pKa = 4.41QNPDD420 pKa = 2.7WQLIQFMIAPTAVFNPFKK438 pKa = 11.04YY439 pKa = 10.22SDD441 pKa = 3.94PQVDD445 pKa = 4.0EE446 pKa = 4.83YY447 pKa = 10.85IQEE450 pKa = 3.82IQYY453 pKa = 11.24GDD455 pKa = 4.11EE456 pKa = 3.94ATQASVAKK464 pKa = 9.68EE465 pKa = 3.68LNTYY469 pKa = 9.78IVEE472 pKa = 4.41QGWFAPFFRR481 pKa = 11.84VQGSVATDD489 pKa = 3.11ANTTVEE495 pKa = 4.19MLPTNAYY502 pKa = 8.08PAIYY506 pKa = 10.15DD507 pKa = 3.79FQPKK511 pKa = 7.63QQ512 pKa = 3.15
Molecular weight: 54.54 kDa
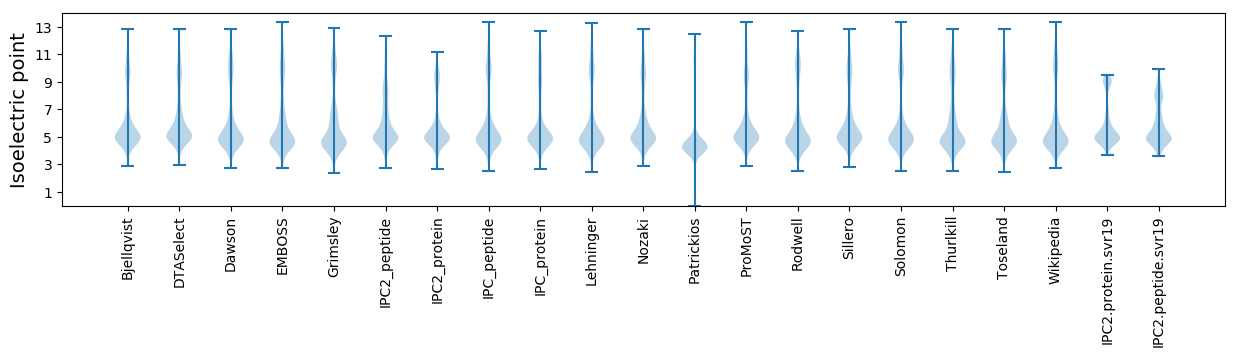
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G7X088|A0A1G7X088_9MICO Demethylmenaquinone methyltransferase OS=Microbacterium pygmaeum OX=370764 GN=menG PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1117582 |
29 |
2029 |
327.5 |
35.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.54 ± 0.053 | 0.476 ± 0.009 |
6.407 ± 0.037 | 5.558 ± 0.039 |
3.182 ± 0.026 | 8.933 ± 0.034 |
1.959 ± 0.021 | 4.827 ± 0.029 |
1.782 ± 0.032 | 10.028 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.788 ± 0.016 | 1.896 ± 0.022 |
5.455 ± 0.036 | 2.756 ± 0.02 |
7.372 ± 0.046 | 5.682 ± 0.031 |
5.982 ± 0.031 | 8.824 ± 0.036 |
1.564 ± 0.018 | 1.99 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
