
Pteropus associated gemycircularvirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus ptero4
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
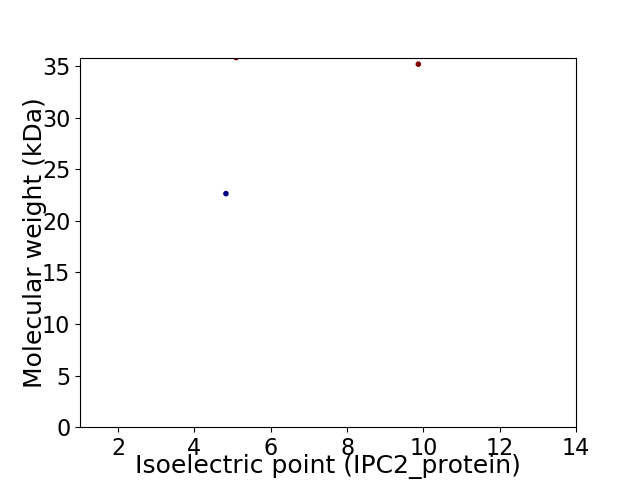
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
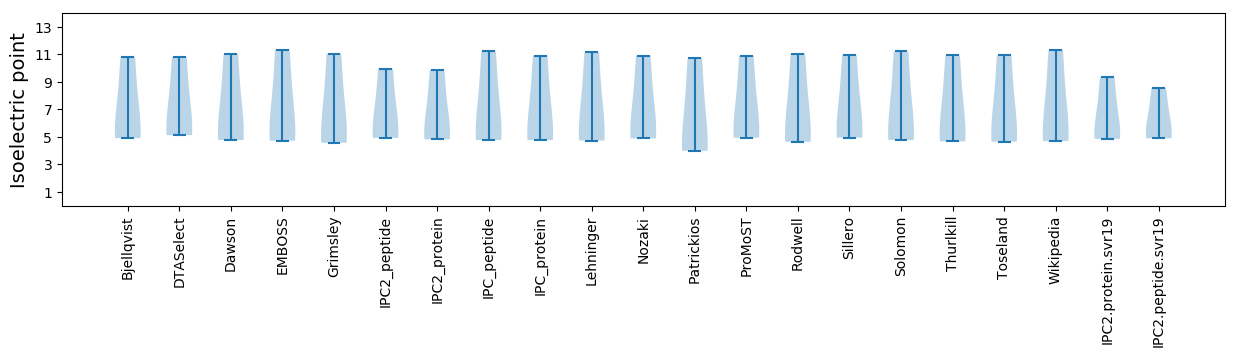
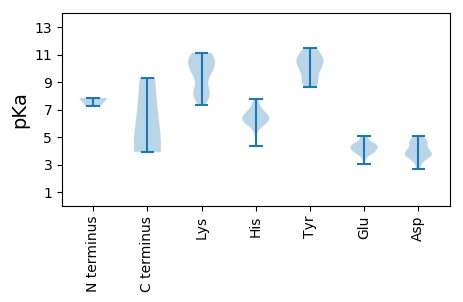
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTS6|A0A140CTS6_9VIRU Replication-associated protein OS=Pteropus associated gemycircularvirus 4 OX=1985398 PE=3 SV=1
MM1 pKa = 7.84PSNGLFVNSRR11 pKa = 11.84YY12 pKa = 10.31KK13 pKa = 10.92LLTYY17 pKa = 8.98AQCGDD22 pKa = 4.68LDD24 pKa = 4.05PFDD27 pKa = 5.0IVDD30 pKa = 4.94LLSGLQGEE38 pKa = 5.05CIVGRR43 pKa = 11.84EE44 pKa = 4.05LHH46 pKa = 6.34SDD48 pKa = 3.64GGIHH52 pKa = 6.48LHH54 pKa = 5.85VFVDD58 pKa = 4.54FGRR61 pKa = 11.84KK62 pKa = 8.03FRR64 pKa = 11.84SRR66 pKa = 11.84SVGIFDD72 pKa = 3.91VGGRR76 pKa = 11.84HH77 pKa = 6.0PNVVASWGTPEE88 pKa = 4.46EE89 pKa = 4.94GYY91 pKa = 10.93DD92 pKa = 3.63YY93 pKa = 10.79AIKK96 pKa = 10.96DD97 pKa = 3.59GDD99 pKa = 4.01VVAGGLEE106 pKa = 3.99RR107 pKa = 11.84PTPRR111 pKa = 11.84EE112 pKa = 3.63SRR114 pKa = 11.84AGNGSAAAKK123 pKa = 8.14WATIADD129 pKa = 3.82ADD131 pKa = 3.94DD132 pKa = 4.26RR133 pKa = 11.84EE134 pKa = 4.42QFFEE138 pKa = 4.51LVKK141 pKa = 11.14SLDD144 pKa = 3.21PKK146 pKa = 10.36TFVTRR151 pKa = 11.84LQDD154 pKa = 3.6LQRR157 pKa = 11.84FADD160 pKa = 3.36WKK162 pKa = 10.48FRR164 pKa = 11.84SDD166 pKa = 3.78PEE168 pKa = 4.52PYY170 pKa = 8.68ITPNNIHH177 pKa = 6.38FPDD180 pKa = 5.06DD181 pKa = 3.8GTDD184 pKa = 4.46GRR186 pKa = 11.84AEE188 pKa = 4.0WVRR191 pKa = 11.84DD192 pKa = 3.54NLVLDD197 pKa = 4.44WEE199 pKa = 4.41NHH201 pKa = 4.33SS202 pKa = 3.92
MM1 pKa = 7.84PSNGLFVNSRR11 pKa = 11.84YY12 pKa = 10.31KK13 pKa = 10.92LLTYY17 pKa = 8.98AQCGDD22 pKa = 4.68LDD24 pKa = 4.05PFDD27 pKa = 5.0IVDD30 pKa = 4.94LLSGLQGEE38 pKa = 5.05CIVGRR43 pKa = 11.84EE44 pKa = 4.05LHH46 pKa = 6.34SDD48 pKa = 3.64GGIHH52 pKa = 6.48LHH54 pKa = 5.85VFVDD58 pKa = 4.54FGRR61 pKa = 11.84KK62 pKa = 8.03FRR64 pKa = 11.84SRR66 pKa = 11.84SVGIFDD72 pKa = 3.91VGGRR76 pKa = 11.84HH77 pKa = 6.0PNVVASWGTPEE88 pKa = 4.46EE89 pKa = 4.94GYY91 pKa = 10.93DD92 pKa = 3.63YY93 pKa = 10.79AIKK96 pKa = 10.96DD97 pKa = 3.59GDD99 pKa = 4.01VVAGGLEE106 pKa = 3.99RR107 pKa = 11.84PTPRR111 pKa = 11.84EE112 pKa = 3.63SRR114 pKa = 11.84AGNGSAAAKK123 pKa = 8.14WATIADD129 pKa = 3.82ADD131 pKa = 3.94DD132 pKa = 4.26RR133 pKa = 11.84EE134 pKa = 4.42QFFEE138 pKa = 4.51LVKK141 pKa = 11.14SLDD144 pKa = 3.21PKK146 pKa = 10.36TFVTRR151 pKa = 11.84LQDD154 pKa = 3.6LQRR157 pKa = 11.84FADD160 pKa = 3.36WKK162 pKa = 10.48FRR164 pKa = 11.84SDD166 pKa = 3.78PEE168 pKa = 4.52PYY170 pKa = 8.68ITPNNIHH177 pKa = 6.38FPDD180 pKa = 5.06DD181 pKa = 3.8GTDD184 pKa = 4.46GRR186 pKa = 11.84AEE188 pKa = 4.0WVRR191 pKa = 11.84DD192 pKa = 3.54NLVLDD197 pKa = 4.44WEE199 pKa = 4.41NHH201 pKa = 4.33SS202 pKa = 3.92
Molecular weight: 22.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTS5|A0A140CTS5_9VIRU RepA OS=Pteropus associated gemycircularvirus 4 OX=1985398 PE=3 SV=1
MM1 pKa = 7.23PRR3 pKa = 11.84YY4 pKa = 8.63YY5 pKa = 11.12AKK7 pKa = 9.51TRR9 pKa = 11.84RR10 pKa = 11.84FRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VIGRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 10.06SSLTGSRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84ITGAKK35 pKa = 8.42RR36 pKa = 11.84RR37 pKa = 11.84SYY39 pKa = 10.34GKK41 pKa = 9.39KK42 pKa = 8.28RR43 pKa = 11.84QMMTTRR49 pKa = 11.84RR50 pKa = 11.84ILNRR54 pKa = 11.84TSTKK58 pKa = 10.36KK59 pKa = 10.23RR60 pKa = 11.84DD61 pKa = 3.58TMLGATTFPPLPEE74 pKa = 4.1NVGALTVTATVPAVLVWCATARR96 pKa = 11.84NLQQEE101 pKa = 4.5ANALDD106 pKa = 4.64TIWSSAQRR114 pKa = 11.84TKK116 pKa = 9.82STCFIRR122 pKa = 11.84GLKK125 pKa = 8.16EE126 pKa = 3.82TITIRR131 pKa = 11.84TNSSAAWRR139 pKa = 11.84WRR141 pKa = 11.84RR142 pKa = 11.84IAFRR146 pKa = 11.84YY147 pKa = 9.73KK148 pKa = 10.36GLLPTGEE155 pKa = 4.06DD156 pKa = 2.86TFARR160 pKa = 11.84FFNEE164 pKa = 3.05ITEE167 pKa = 4.26ANRR170 pKa = 11.84VRR172 pKa = 11.84IMRR175 pKa = 11.84PVTALPFSIVSPLYY189 pKa = 9.2AHH191 pKa = 7.75IFRR194 pKa = 11.84GFGINDD200 pKa = 4.04LSANPQDD207 pKa = 3.4WVDD210 pKa = 4.82PITAPVDD217 pKa = 3.43TDD219 pKa = 4.83RR220 pKa = 11.84IAPLYY225 pKa = 10.3DD226 pKa = 2.77KK227 pKa = 10.8VVRR230 pKa = 11.84ITSGNDD236 pKa = 2.67AGVANTYY243 pKa = 10.2RR244 pKa = 11.84IWHH247 pKa = 6.23PVNKK251 pKa = 9.73NLRR254 pKa = 11.84YY255 pKa = 10.02NDD257 pKa = 3.69TEE259 pKa = 4.2FGGGFSSSPLSTEE272 pKa = 3.8GRR274 pKa = 11.84PGGGDD279 pKa = 3.64FYY281 pKa = 11.42VIDD284 pKa = 4.77IIIGNSSDD292 pKa = 4.51PGDD295 pKa = 4.06LLDD298 pKa = 4.35WLPTSTLYY306 pKa = 9.73WHH308 pKa = 7.06EE309 pKa = 4.2KK310 pKa = 9.32
MM1 pKa = 7.23PRR3 pKa = 11.84YY4 pKa = 8.63YY5 pKa = 11.12AKK7 pKa = 9.51TRR9 pKa = 11.84RR10 pKa = 11.84FRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VIGRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 10.06SSLTGSRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84ITGAKK35 pKa = 8.42RR36 pKa = 11.84RR37 pKa = 11.84SYY39 pKa = 10.34GKK41 pKa = 9.39KK42 pKa = 8.28RR43 pKa = 11.84QMMTTRR49 pKa = 11.84RR50 pKa = 11.84ILNRR54 pKa = 11.84TSTKK58 pKa = 10.36KK59 pKa = 10.23RR60 pKa = 11.84DD61 pKa = 3.58TMLGATTFPPLPEE74 pKa = 4.1NVGALTVTATVPAVLVWCATARR96 pKa = 11.84NLQQEE101 pKa = 4.5ANALDD106 pKa = 4.64TIWSSAQRR114 pKa = 11.84TKK116 pKa = 9.82STCFIRR122 pKa = 11.84GLKK125 pKa = 8.16EE126 pKa = 3.82TITIRR131 pKa = 11.84TNSSAAWRR139 pKa = 11.84WRR141 pKa = 11.84RR142 pKa = 11.84IAFRR146 pKa = 11.84YY147 pKa = 9.73KK148 pKa = 10.36GLLPTGEE155 pKa = 4.06DD156 pKa = 2.86TFARR160 pKa = 11.84FFNEE164 pKa = 3.05ITEE167 pKa = 4.26ANRR170 pKa = 11.84VRR172 pKa = 11.84IMRR175 pKa = 11.84PVTALPFSIVSPLYY189 pKa = 9.2AHH191 pKa = 7.75IFRR194 pKa = 11.84GFGINDD200 pKa = 4.04LSANPQDD207 pKa = 3.4WVDD210 pKa = 4.82PITAPVDD217 pKa = 3.43TDD219 pKa = 4.83RR220 pKa = 11.84IAPLYY225 pKa = 10.3DD226 pKa = 2.77KK227 pKa = 10.8VVRR230 pKa = 11.84ITSGNDD236 pKa = 2.67AGVANTYY243 pKa = 10.2RR244 pKa = 11.84IWHH247 pKa = 6.23PVNKK251 pKa = 9.73NLRR254 pKa = 11.84YY255 pKa = 10.02NDD257 pKa = 3.69TEE259 pKa = 4.2FGGGFSSSPLSTEE272 pKa = 3.8GRR274 pKa = 11.84PGGGDD279 pKa = 3.64FYY281 pKa = 11.42VIDD284 pKa = 4.77IIIGNSSDD292 pKa = 4.51PGDD295 pKa = 4.06LLDD298 pKa = 4.35WLPTSTLYY306 pKa = 9.73WHH308 pKa = 7.06EE309 pKa = 4.2KK310 pKa = 9.32
Molecular weight: 35.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
829 |
202 |
317 |
276.3 |
31.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.393 ± 0.561 | 1.327 ± 0.442 |
8.203 ± 1.553 | 4.101 ± 0.652 |
5.549 ± 0.682 | 8.806 ± 0.843 |
2.292 ± 0.652 | 5.066 ± 0.843 |
4.101 ± 0.242 | 7.358 ± 0.304 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.206 ± 0.282 | 4.584 ± 0.233 |
5.187 ± 0.39 | 2.533 ± 0.507 |
8.444 ± 1.756 | 6.393 ± 0.212 |
6.273 ± 1.993 | 6.514 ± 0.669 |
2.774 ± 0.19 | 2.895 ± 0.189 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
