
Cellulomonas marina
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Cellulomonadaceae; Cellulomonas
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
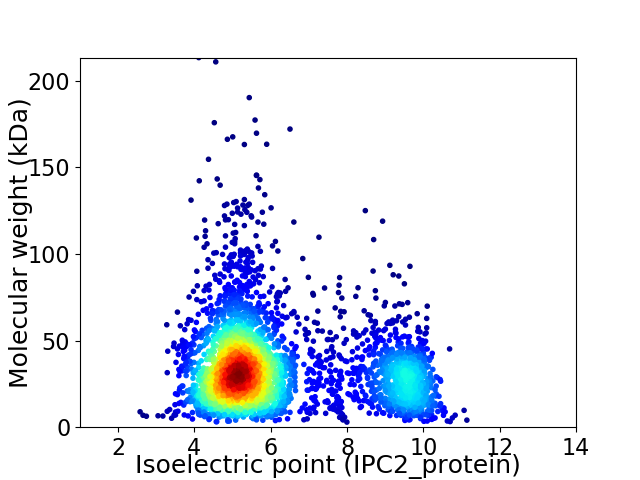
Virtual 2D-PAGE plot for 3460 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
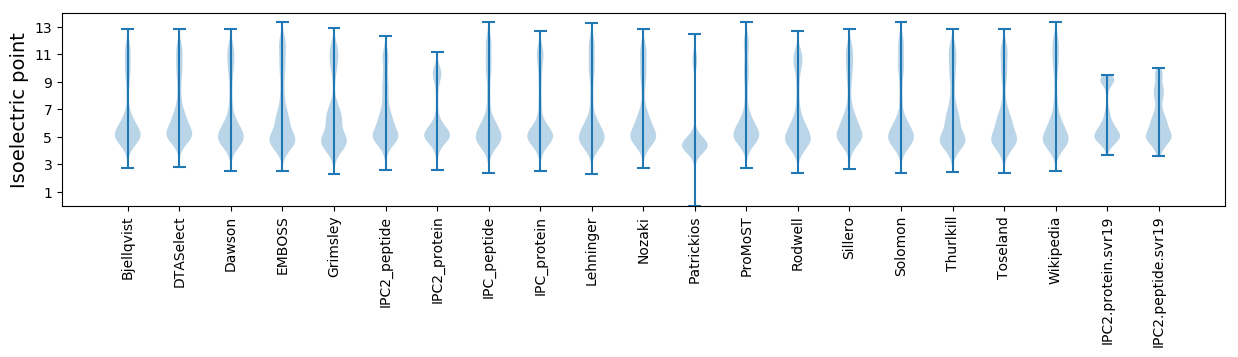
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I0VWE3|A0A1I0VWE3_9CELL Deoxyguanosinetriphosphate triphosphohydrolase-like protein OS=Cellulomonas marina OX=988821 GN=SAMN05421867_1026 PE=3 SV=1
MM1 pKa = 7.54RR2 pKa = 11.84RR3 pKa = 11.84ITSTVAMMAGVTLLAAACSGGGGSTSAEE31 pKa = 4.01EE32 pKa = 4.53SPSSSAAAEE41 pKa = 4.07PSASEE46 pKa = 3.7SAPPVRR52 pKa = 11.84GDD54 pKa = 3.45EE55 pKa = 4.27EE56 pKa = 4.42LVIWTDD62 pKa = 3.26ALKK65 pKa = 10.74LDD67 pKa = 4.01AVRR70 pKa = 11.84AVADD74 pKa = 4.0QFAEE78 pKa = 4.4DD79 pKa = 3.34NGITVGVQAIVNTRR93 pKa = 11.84TDD95 pKa = 4.61FITANAAGNGPDD107 pKa = 3.73VVVGAHH113 pKa = 6.32DD114 pKa = 4.31WIGQLVQNGAIDD126 pKa = 4.17PLQLSAADD134 pKa = 3.58LGGYY138 pKa = 9.69AEE140 pKa = 4.35NAIQAVTYY148 pKa = 10.18DD149 pKa = 3.52GQIYY153 pKa = 8.83GLPYY157 pKa = 10.14GVEE160 pKa = 4.19ALGLYY165 pKa = 9.65CNKK168 pKa = 10.25AYY170 pKa = 10.42APDD173 pKa = 4.16TYY175 pKa = 11.55ASLDD179 pKa = 3.66DD180 pKa = 4.73VIAAGQAAVDD190 pKa = 3.93AGQVEE195 pKa = 4.59TSLNVAQGDD204 pKa = 3.87VGDD207 pKa = 4.78AYY209 pKa = 11.03HH210 pKa = 6.15MQPILTSMGGYY221 pKa = 10.64LFGQNPDD228 pKa = 2.88GTYY231 pKa = 10.68NPEE234 pKa = 4.24DD235 pKa = 4.26LGLASPGGLAAAQKK249 pKa = 10.37IFDD252 pKa = 4.55LGEE255 pKa = 3.83QGSNVLRR262 pKa = 11.84RR263 pKa = 11.84SISGDD268 pKa = 2.89NSIALFAEE276 pKa = 4.75GRR278 pKa = 11.84AACLISGPWALADD291 pKa = 3.48VRR293 pKa = 11.84EE294 pKa = 4.25GLGEE298 pKa = 4.59DD299 pKa = 5.04GYY301 pKa = 9.33TLQPIPGFAGQQPAQPFLGAQAFYY325 pKa = 11.1VAANGQNKK333 pKa = 7.95TFAQEE338 pKa = 3.93FVTNGVNNAEE348 pKa = 4.49GMTTMYY354 pKa = 10.51EE355 pKa = 4.15LANLPPALTEE365 pKa = 4.0VRR367 pKa = 11.84QSIAADD373 pKa = 3.42NPDD376 pKa = 3.61FEE378 pKa = 5.38VFAQAADD385 pKa = 3.65NGNPMPAVPAMAEE398 pKa = 3.68VWAPLGQAYY407 pKa = 10.14SAIIGGADD415 pKa = 3.58PASTMQQAQDD425 pKa = 3.79TIAAAIAASS434 pKa = 3.48
MM1 pKa = 7.54RR2 pKa = 11.84RR3 pKa = 11.84ITSTVAMMAGVTLLAAACSGGGGSTSAEE31 pKa = 4.01EE32 pKa = 4.53SPSSSAAAEE41 pKa = 4.07PSASEE46 pKa = 3.7SAPPVRR52 pKa = 11.84GDD54 pKa = 3.45EE55 pKa = 4.27EE56 pKa = 4.42LVIWTDD62 pKa = 3.26ALKK65 pKa = 10.74LDD67 pKa = 4.01AVRR70 pKa = 11.84AVADD74 pKa = 4.0QFAEE78 pKa = 4.4DD79 pKa = 3.34NGITVGVQAIVNTRR93 pKa = 11.84TDD95 pKa = 4.61FITANAAGNGPDD107 pKa = 3.73VVVGAHH113 pKa = 6.32DD114 pKa = 4.31WIGQLVQNGAIDD126 pKa = 4.17PLQLSAADD134 pKa = 3.58LGGYY138 pKa = 9.69AEE140 pKa = 4.35NAIQAVTYY148 pKa = 10.18DD149 pKa = 3.52GQIYY153 pKa = 8.83GLPYY157 pKa = 10.14GVEE160 pKa = 4.19ALGLYY165 pKa = 9.65CNKK168 pKa = 10.25AYY170 pKa = 10.42APDD173 pKa = 4.16TYY175 pKa = 11.55ASLDD179 pKa = 3.66DD180 pKa = 4.73VIAAGQAAVDD190 pKa = 3.93AGQVEE195 pKa = 4.59TSLNVAQGDD204 pKa = 3.87VGDD207 pKa = 4.78AYY209 pKa = 11.03HH210 pKa = 6.15MQPILTSMGGYY221 pKa = 10.64LFGQNPDD228 pKa = 2.88GTYY231 pKa = 10.68NPEE234 pKa = 4.24DD235 pKa = 4.26LGLASPGGLAAAQKK249 pKa = 10.37IFDD252 pKa = 4.55LGEE255 pKa = 3.83QGSNVLRR262 pKa = 11.84RR263 pKa = 11.84SISGDD268 pKa = 2.89NSIALFAEE276 pKa = 4.75GRR278 pKa = 11.84AACLISGPWALADD291 pKa = 3.48VRR293 pKa = 11.84EE294 pKa = 4.25GLGEE298 pKa = 4.59DD299 pKa = 5.04GYY301 pKa = 9.33TLQPIPGFAGQQPAQPFLGAQAFYY325 pKa = 11.1VAANGQNKK333 pKa = 7.95TFAQEE338 pKa = 3.93FVTNGVNNAEE348 pKa = 4.49GMTTMYY354 pKa = 10.51EE355 pKa = 4.15LANLPPALTEE365 pKa = 4.0VRR367 pKa = 11.84QSIAADD373 pKa = 3.42NPDD376 pKa = 3.61FEE378 pKa = 5.38VFAQAADD385 pKa = 3.65NGNPMPAVPAMAEE398 pKa = 3.68VWAPLGQAYY407 pKa = 10.14SAIIGGADD415 pKa = 3.58PASTMQQAQDD425 pKa = 3.79TIAAAIAASS434 pKa = 3.48
Molecular weight: 44.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I0XJB0|A0A1I0XJB0_9CELL 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase OS=Cellulomonas marina OX=988821 GN=menD PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1193049 |
30 |
2100 |
344.8 |
36.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.438 ± 0.07 | 0.535 ± 0.011 |
6.375 ± 0.031 | 5.198 ± 0.039 |
2.227 ± 0.025 | 9.9 ± 0.045 |
2.095 ± 0.019 | 2.104 ± 0.029 |
1.113 ± 0.021 | 10.608 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.355 ± 0.017 | 1.224 ± 0.018 |
6.364 ± 0.042 | 2.501 ± 0.023 |
8.361 ± 0.043 | 4.462 ± 0.03 |
6.347 ± 0.042 | 10.65 ± 0.047 |
1.505 ± 0.018 | 1.638 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
