
Cochleicola gelatinilyticus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Cochleicola
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
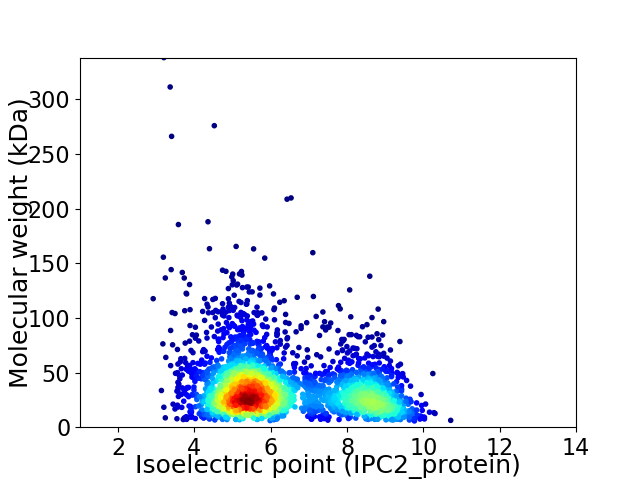
Virtual 2D-PAGE plot for 2962 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A167G804|A0A167G804_9FLAO tRNA modification GTPase MnmE OS=Cochleicola gelatinilyticus OX=1763537 GN=mnmE PE=3 SV=1
MM1 pKa = 7.61KK2 pKa = 10.4KK3 pKa = 10.27LLYY6 pKa = 10.56LFVSGLILASCSVDD20 pKa = 3.31QPEE23 pKa = 4.39TSEE26 pKa = 4.42SNADD30 pKa = 3.64LAYY33 pKa = 10.49DD34 pKa = 4.46ASTLDD39 pKa = 3.63ASSLGNYY46 pKa = 9.35KK47 pKa = 10.64GVFTTNNSDD56 pKa = 3.05YY57 pKa = 10.46RR58 pKa = 11.84ATVEE62 pKa = 3.88IDD64 pKa = 3.24VPYY67 pKa = 10.39FNSEE71 pKa = 3.65NTTKK75 pKa = 10.52QKK77 pKa = 10.73SYY79 pKa = 8.8PTALVSLQNGKK90 pKa = 10.09SFVATANRR98 pKa = 11.84NISLGEE104 pKa = 4.29EE105 pKa = 3.7ITKK108 pKa = 10.73LEE110 pKa = 4.12FSGSEE115 pKa = 3.69MSFKK119 pKa = 10.77FSVNDD124 pKa = 4.34DD125 pKa = 3.64GTSPIITDD133 pKa = 3.33VNYY136 pKa = 10.85KK137 pKa = 9.35NLKK140 pKa = 10.01GDD142 pKa = 3.42ILVAKK147 pKa = 7.87NTSFAPVTTFTGTYY161 pKa = 9.93QCFQCGNSEE170 pKa = 3.85PRR172 pKa = 11.84TWNVVISGDD181 pKa = 3.37ATGDD185 pKa = 3.24QTYY188 pKa = 8.26ATQVEE193 pKa = 5.03FNGNTYY199 pKa = 10.78SGGQGLQEE207 pKa = 4.01NCVVDD212 pKa = 4.01GTNSDD217 pKa = 3.18ITFCTSRR224 pKa = 11.84SGDD227 pKa = 3.18AGSNVGFMVGSNAVEE242 pKa = 4.15WEE244 pKa = 4.01GQMGYY249 pKa = 10.84SNSAPDD255 pKa = 4.0CSSQVGFWYY264 pKa = 9.82FKK266 pKa = 10.68RR267 pKa = 11.84GLEE270 pKa = 4.18GQKK273 pKa = 10.51SGTFKK278 pKa = 10.45TDD280 pKa = 2.43AMNPEE285 pKa = 4.23SPNCFTRR292 pKa = 11.84LAFEE296 pKa = 4.25NFEE299 pKa = 5.46DD300 pKa = 3.92DD301 pKa = 3.27TLMYY305 pKa = 7.42TTSEE309 pKa = 4.34PEE311 pKa = 4.04FTSGNDD317 pKa = 3.37DD318 pKa = 3.58YY319 pKa = 11.11FTRR322 pKa = 11.84TDD324 pKa = 3.25GSNIGASFNNFSNLIGNGYY343 pKa = 9.23FAAQDD348 pKa = 3.25VDD350 pKa = 4.06SQIGIPAYY358 pKa = 10.02IMFEE362 pKa = 4.3NINIASFSTVYY373 pKa = 10.68VDD375 pKa = 5.22ASFAEE380 pKa = 4.87DD381 pKa = 3.5DD382 pKa = 4.26ASNNDD387 pKa = 3.78EE388 pKa = 5.32DD389 pKa = 4.84WDD391 pKa = 3.93NTDD394 pKa = 3.33NVIMEE399 pKa = 4.01YY400 pKa = 10.81SFDD403 pKa = 3.78GTNWTPILAITNDD416 pKa = 3.17GSQFNSAPQIDD427 pKa = 3.95TDD429 pKa = 3.97FDD431 pKa = 4.22GVGDD435 pKa = 3.85GVIITEE441 pKa = 4.53TFQDD445 pKa = 3.47FRR447 pKa = 11.84VAFNNNVANNPTASGTVAIRR467 pKa = 11.84IAINLNAGDD476 pKa = 4.12EE477 pKa = 4.88DD478 pKa = 3.85IAIDD482 pKa = 3.89NILIRR487 pKa = 11.84GYY489 pKa = 11.05
MM1 pKa = 7.61KK2 pKa = 10.4KK3 pKa = 10.27LLYY6 pKa = 10.56LFVSGLILASCSVDD20 pKa = 3.31QPEE23 pKa = 4.39TSEE26 pKa = 4.42SNADD30 pKa = 3.64LAYY33 pKa = 10.49DD34 pKa = 4.46ASTLDD39 pKa = 3.63ASSLGNYY46 pKa = 9.35KK47 pKa = 10.64GVFTTNNSDD56 pKa = 3.05YY57 pKa = 10.46RR58 pKa = 11.84ATVEE62 pKa = 3.88IDD64 pKa = 3.24VPYY67 pKa = 10.39FNSEE71 pKa = 3.65NTTKK75 pKa = 10.52QKK77 pKa = 10.73SYY79 pKa = 8.8PTALVSLQNGKK90 pKa = 10.09SFVATANRR98 pKa = 11.84NISLGEE104 pKa = 4.29EE105 pKa = 3.7ITKK108 pKa = 10.73LEE110 pKa = 4.12FSGSEE115 pKa = 3.69MSFKK119 pKa = 10.77FSVNDD124 pKa = 4.34DD125 pKa = 3.64GTSPIITDD133 pKa = 3.33VNYY136 pKa = 10.85KK137 pKa = 9.35NLKK140 pKa = 10.01GDD142 pKa = 3.42ILVAKK147 pKa = 7.87NTSFAPVTTFTGTYY161 pKa = 9.93QCFQCGNSEE170 pKa = 3.85PRR172 pKa = 11.84TWNVVISGDD181 pKa = 3.37ATGDD185 pKa = 3.24QTYY188 pKa = 8.26ATQVEE193 pKa = 5.03FNGNTYY199 pKa = 10.78SGGQGLQEE207 pKa = 4.01NCVVDD212 pKa = 4.01GTNSDD217 pKa = 3.18ITFCTSRR224 pKa = 11.84SGDD227 pKa = 3.18AGSNVGFMVGSNAVEE242 pKa = 4.15WEE244 pKa = 4.01GQMGYY249 pKa = 10.84SNSAPDD255 pKa = 4.0CSSQVGFWYY264 pKa = 9.82FKK266 pKa = 10.68RR267 pKa = 11.84GLEE270 pKa = 4.18GQKK273 pKa = 10.51SGTFKK278 pKa = 10.45TDD280 pKa = 2.43AMNPEE285 pKa = 4.23SPNCFTRR292 pKa = 11.84LAFEE296 pKa = 4.25NFEE299 pKa = 5.46DD300 pKa = 3.92DD301 pKa = 3.27TLMYY305 pKa = 7.42TTSEE309 pKa = 4.34PEE311 pKa = 4.04FTSGNDD317 pKa = 3.37DD318 pKa = 3.58YY319 pKa = 11.11FTRR322 pKa = 11.84TDD324 pKa = 3.25GSNIGASFNNFSNLIGNGYY343 pKa = 9.23FAAQDD348 pKa = 3.25VDD350 pKa = 4.06SQIGIPAYY358 pKa = 10.02IMFEE362 pKa = 4.3NINIASFSTVYY373 pKa = 10.68VDD375 pKa = 5.22ASFAEE380 pKa = 4.87DD381 pKa = 3.5DD382 pKa = 4.26ASNNDD387 pKa = 3.78EE388 pKa = 5.32DD389 pKa = 4.84WDD391 pKa = 3.93NTDD394 pKa = 3.33NVIMEE399 pKa = 4.01YY400 pKa = 10.81SFDD403 pKa = 3.78GTNWTPILAITNDD416 pKa = 3.17GSQFNSAPQIDD427 pKa = 3.95TDD429 pKa = 3.97FDD431 pKa = 4.22GVGDD435 pKa = 3.85GVIITEE441 pKa = 4.53TFQDD445 pKa = 3.47FRR447 pKa = 11.84VAFNNNVANNPTASGTVAIRR467 pKa = 11.84IAINLNAGDD476 pKa = 4.12EE477 pKa = 4.88DD478 pKa = 3.85IAIDD482 pKa = 3.89NILIRR487 pKa = 11.84GYY489 pKa = 11.05
Molecular weight: 53.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A167H472|A0A167H472_9FLAO tRNA (N(6)-L-threonylcarbamoyladenosine(37)-C(2))-methylthiotransferase MtaB OS=Cochleicola gelatinilyticus OX=1763537 GN=ULVI_12020 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.17VLAARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.87GRR40 pKa = 11.84KK41 pKa = 8.56KK42 pKa = 10.21ISVSSEE48 pKa = 3.74TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.17VLAARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.87GRR40 pKa = 11.84KK41 pKa = 8.56KK42 pKa = 10.21ISVSSEE48 pKa = 3.74TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1004104 |
50 |
3207 |
339.0 |
38.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.678 ± 0.033 | 0.714 ± 0.016 |
5.514 ± 0.034 | 6.986 ± 0.044 |
5.202 ± 0.037 | 6.527 ± 0.059 |
1.756 ± 0.022 | 7.589 ± 0.039 |
7.268 ± 0.069 | 9.208 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.154 ± 0.022 | 5.895 ± 0.046 |
3.47 ± 0.03 | 3.418 ± 0.028 |
3.594 ± 0.031 | 6.492 ± 0.033 |
6.309 ± 0.05 | 6.314 ± 0.035 |
0.977 ± 0.016 | 3.937 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
