
Staphylococcus phage StB20
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
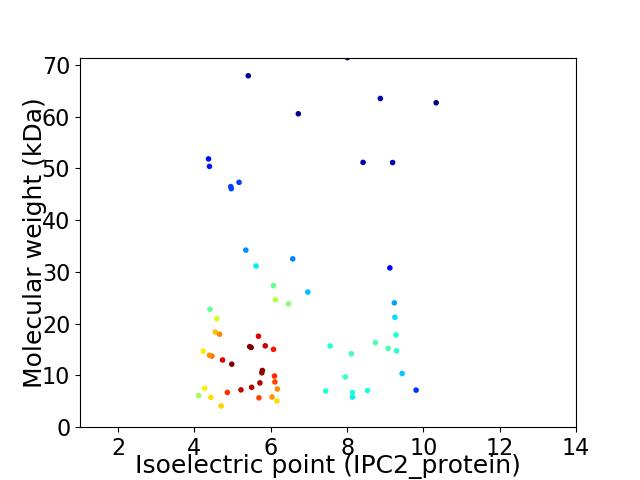
Virtual 2D-PAGE plot for 65 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|H9A136|H9A136_9CAUD Uncharacterized protein OS=Staphylococcus phage StB20 OX=1147043 PE=4 SV=1
MM1 pKa = 7.48IIKK4 pKa = 9.28MGEE7 pKa = 4.13VEE9 pKa = 4.1THH11 pKa = 5.32INPRR15 pKa = 11.84NVDD18 pKa = 2.84IGDD21 pKa = 3.47IGSRR25 pKa = 11.84FYY27 pKa = 11.1TEE29 pKa = 4.84DD30 pKa = 3.4EE31 pKa = 3.81NTAFIRR37 pKa = 11.84IRR39 pKa = 11.84INYY42 pKa = 8.26NGSPVDD48 pKa = 3.83LTKK51 pKa = 10.78TDD53 pKa = 4.04MKK55 pKa = 10.73PKK57 pKa = 10.28LDD59 pKa = 4.12LFMEE63 pKa = 5.06DD64 pKa = 2.93GSVFIDD70 pKa = 3.4EE71 pKa = 4.83TIEE74 pKa = 3.88VLIPEE79 pKa = 4.76IGLIKK84 pKa = 10.39YY85 pKa = 7.56DD86 pKa = 3.39IPSNVIKK93 pKa = 10.76HH94 pKa = 5.58IGKK97 pKa = 9.58VNCKK101 pKa = 10.24LFMDD105 pKa = 4.14SLEE108 pKa = 4.64HH109 pKa = 6.13SVHH112 pKa = 5.37VANFSFTIVDD122 pKa = 3.71SGVEE126 pKa = 3.71EE127 pKa = 4.33AVAKK131 pKa = 9.68EE132 pKa = 3.88VNLNIVKK139 pKa = 9.1DD140 pKa = 3.84TVKK143 pKa = 10.76QIMSEE148 pKa = 4.09DD149 pKa = 3.76LVDD152 pKa = 4.66LLNDD156 pKa = 3.75GFKK159 pKa = 11.11DD160 pKa = 3.39KK161 pKa = 10.67LTGDD165 pKa = 4.03LEE167 pKa = 4.89NYY169 pKa = 10.29LSTHH173 pKa = 5.32NEE175 pKa = 3.68EE176 pKa = 4.48FKK178 pKa = 11.15GPKK181 pKa = 8.91GDD183 pKa = 4.28KK184 pKa = 10.59GPQGEE189 pKa = 4.41QGPQGLQGEE198 pKa = 4.34QGIQGEE204 pKa = 4.55KK205 pKa = 10.64GEE207 pKa = 4.51QGPQGATGPQGPPGKK222 pKa = 9.91DD223 pKa = 2.99GRR225 pKa = 11.84DD226 pKa = 3.61GVDD229 pKa = 3.2GKK231 pKa = 11.42SFAYY235 pKa = 10.31DD236 pKa = 3.42DD237 pKa = 4.07FTPEE241 pKa = 4.13QLDD244 pKa = 3.49KK245 pKa = 11.32LKK247 pKa = 10.9PEE249 pKa = 4.82LPDD252 pKa = 3.74FSKK255 pKa = 8.78WQKK258 pKa = 11.08YY259 pKa = 9.85KK260 pKa = 10.09LTNDD264 pKa = 4.49DD265 pKa = 3.88GNQIATFDD273 pKa = 3.58SDD275 pKa = 4.19NRR277 pKa = 11.84MTNEE281 pKa = 4.13NLLSTPTGFYY291 pKa = 11.0YY292 pKa = 10.54NYY294 pKa = 10.2EE295 pKa = 4.09YY296 pKa = 10.9DD297 pKa = 3.88DD298 pKa = 4.53SPNGSEE304 pKa = 3.88GVVYY308 pKa = 10.94VMEE311 pKa = 4.3YY312 pKa = 10.21TDD314 pKa = 3.53EE315 pKa = 4.3CKK317 pKa = 10.21KK318 pKa = 10.85VYY320 pKa = 9.99YY321 pKa = 10.13SSYY324 pKa = 10.36RR325 pKa = 11.84YY326 pKa = 9.94NDD328 pKa = 4.11LYY330 pKa = 11.42VKK332 pKa = 9.67TYY334 pKa = 10.57HH335 pKa = 7.45PDD337 pKa = 3.54FGWLEE342 pKa = 3.93WEE344 pKa = 4.74EE345 pKa = 3.94ISNGYY350 pKa = 7.89TDD352 pKa = 4.0TGWIPLAIKK361 pKa = 10.37NDD363 pKa = 3.62YY364 pKa = 10.77EE365 pKa = 5.27IITNLNYY372 pKa = 8.77EE373 pKa = 3.61QSYY376 pKa = 9.94RR377 pKa = 11.84VIDD380 pKa = 3.43YY381 pKa = 11.49GNFKK385 pKa = 10.17KK386 pKa = 11.05VSVRR390 pKa = 11.84LGVIDD395 pKa = 4.32NSPSNHH401 pKa = 5.79SVVASIPEE409 pKa = 4.0YY410 pKa = 10.77LNPYY414 pKa = 10.17DD415 pKa = 3.53VFGYY419 pKa = 10.42GVSTTQGGRR428 pKa = 11.84PVTVTIVDD436 pKa = 3.97GDD438 pKa = 3.46IMLYY442 pKa = 10.41SNSFASSMNTYY453 pKa = 9.9YY454 pKa = 10.37QGEE457 pKa = 4.34WIII460 pKa = 4.24
MM1 pKa = 7.48IIKK4 pKa = 9.28MGEE7 pKa = 4.13VEE9 pKa = 4.1THH11 pKa = 5.32INPRR15 pKa = 11.84NVDD18 pKa = 2.84IGDD21 pKa = 3.47IGSRR25 pKa = 11.84FYY27 pKa = 11.1TEE29 pKa = 4.84DD30 pKa = 3.4EE31 pKa = 3.81NTAFIRR37 pKa = 11.84IRR39 pKa = 11.84INYY42 pKa = 8.26NGSPVDD48 pKa = 3.83LTKK51 pKa = 10.78TDD53 pKa = 4.04MKK55 pKa = 10.73PKK57 pKa = 10.28LDD59 pKa = 4.12LFMEE63 pKa = 5.06DD64 pKa = 2.93GSVFIDD70 pKa = 3.4EE71 pKa = 4.83TIEE74 pKa = 3.88VLIPEE79 pKa = 4.76IGLIKK84 pKa = 10.39YY85 pKa = 7.56DD86 pKa = 3.39IPSNVIKK93 pKa = 10.76HH94 pKa = 5.58IGKK97 pKa = 9.58VNCKK101 pKa = 10.24LFMDD105 pKa = 4.14SLEE108 pKa = 4.64HH109 pKa = 6.13SVHH112 pKa = 5.37VANFSFTIVDD122 pKa = 3.71SGVEE126 pKa = 3.71EE127 pKa = 4.33AVAKK131 pKa = 9.68EE132 pKa = 3.88VNLNIVKK139 pKa = 9.1DD140 pKa = 3.84TVKK143 pKa = 10.76QIMSEE148 pKa = 4.09DD149 pKa = 3.76LVDD152 pKa = 4.66LLNDD156 pKa = 3.75GFKK159 pKa = 11.11DD160 pKa = 3.39KK161 pKa = 10.67LTGDD165 pKa = 4.03LEE167 pKa = 4.89NYY169 pKa = 10.29LSTHH173 pKa = 5.32NEE175 pKa = 3.68EE176 pKa = 4.48FKK178 pKa = 11.15GPKK181 pKa = 8.91GDD183 pKa = 4.28KK184 pKa = 10.59GPQGEE189 pKa = 4.41QGPQGLQGEE198 pKa = 4.34QGIQGEE204 pKa = 4.55KK205 pKa = 10.64GEE207 pKa = 4.51QGPQGATGPQGPPGKK222 pKa = 9.91DD223 pKa = 2.99GRR225 pKa = 11.84DD226 pKa = 3.61GVDD229 pKa = 3.2GKK231 pKa = 11.42SFAYY235 pKa = 10.31DD236 pKa = 3.42DD237 pKa = 4.07FTPEE241 pKa = 4.13QLDD244 pKa = 3.49KK245 pKa = 11.32LKK247 pKa = 10.9PEE249 pKa = 4.82LPDD252 pKa = 3.74FSKK255 pKa = 8.78WQKK258 pKa = 11.08YY259 pKa = 9.85KK260 pKa = 10.09LTNDD264 pKa = 4.49DD265 pKa = 3.88GNQIATFDD273 pKa = 3.58SDD275 pKa = 4.19NRR277 pKa = 11.84MTNEE281 pKa = 4.13NLLSTPTGFYY291 pKa = 11.0YY292 pKa = 10.54NYY294 pKa = 10.2EE295 pKa = 4.09YY296 pKa = 10.9DD297 pKa = 3.88DD298 pKa = 4.53SPNGSEE304 pKa = 3.88GVVYY308 pKa = 10.94VMEE311 pKa = 4.3YY312 pKa = 10.21TDD314 pKa = 3.53EE315 pKa = 4.3CKK317 pKa = 10.21KK318 pKa = 10.85VYY320 pKa = 9.99YY321 pKa = 10.13SSYY324 pKa = 10.36RR325 pKa = 11.84YY326 pKa = 9.94NDD328 pKa = 4.11LYY330 pKa = 11.42VKK332 pKa = 9.67TYY334 pKa = 10.57HH335 pKa = 7.45PDD337 pKa = 3.54FGWLEE342 pKa = 3.93WEE344 pKa = 4.74EE345 pKa = 3.94ISNGYY350 pKa = 7.89TDD352 pKa = 4.0TGWIPLAIKK361 pKa = 10.37NDD363 pKa = 3.62YY364 pKa = 10.77EE365 pKa = 5.27IITNLNYY372 pKa = 8.77EE373 pKa = 3.61QSYY376 pKa = 9.94RR377 pKa = 11.84VIDD380 pKa = 3.43YY381 pKa = 11.49GNFKK385 pKa = 10.17KK386 pKa = 11.05VSVRR390 pKa = 11.84LGVIDD395 pKa = 4.32NSPSNHH401 pKa = 5.79SVVASIPEE409 pKa = 4.0YY410 pKa = 10.77LNPYY414 pKa = 10.17DD415 pKa = 3.53VFGYY419 pKa = 10.42GVSTTQGGRR428 pKa = 11.84PVTVTIVDD436 pKa = 3.97GDD438 pKa = 3.46IMLYY442 pKa = 10.41SNSFASSMNTYY453 pKa = 9.9YY454 pKa = 10.37QGEE457 pKa = 4.34WIII460 pKa = 4.24
Molecular weight: 51.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H9A102|H9A102_9CAUD Uncharacterized protein OS=Staphylococcus phage StB20 OX=1147043 PE=4 SV=1
MM1 pKa = 7.45ILSKK5 pKa = 9.87TINQRR10 pKa = 11.84YY11 pKa = 9.27KK12 pKa = 11.01YY13 pKa = 8.05NTQGKK18 pKa = 7.18TPTEE22 pKa = 4.06VQRR25 pKa = 11.84EE26 pKa = 3.98LRR28 pKa = 11.84QLGVKK33 pKa = 10.18GFVVKK38 pKa = 10.62VAGNRR43 pKa = 11.84VTMKK47 pKa = 10.57VNEE50 pKa = 3.91NDD52 pKa = 2.91IKK54 pKa = 11.02KK55 pKa = 9.99NRR57 pKa = 11.84EE58 pKa = 3.66FLRR61 pKa = 4.98
MM1 pKa = 7.45ILSKK5 pKa = 9.87TINQRR10 pKa = 11.84YY11 pKa = 9.27KK12 pKa = 11.01YY13 pKa = 8.05NTQGKK18 pKa = 7.18TPTEE22 pKa = 4.06VQRR25 pKa = 11.84EE26 pKa = 3.98LRR28 pKa = 11.84QLGVKK33 pKa = 10.18GFVVKK38 pKa = 10.62VAGNRR43 pKa = 11.84VTMKK47 pKa = 10.57VNEE50 pKa = 3.91NDD52 pKa = 2.91IKK54 pKa = 11.02KK55 pKa = 9.99NRR57 pKa = 11.84EE58 pKa = 3.66FLRR61 pKa = 4.98
Molecular weight: 7.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12593 |
34 |
620 |
193.7 |
22.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.289 ± 0.497 | 0.604 ± 0.099 |
6.313 ± 0.375 | 7.52 ± 0.508 |
3.947 ± 0.224 | 6.218 ± 0.506 |
1.803 ± 0.181 | 7.25 ± 0.328 |
9.426 ± 0.42 | 7.552 ± 0.253 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.74 ± 0.193 | 6.885 ± 0.365 |
3.121 ± 0.226 | 3.748 ± 0.212 |
4.066 ± 0.256 | 5.892 ± 0.282 |
6.003 ± 0.236 | 6.194 ± 0.174 |
1.247 ± 0.133 | 4.185 ± 0.29 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
