
Mulberry vein banding virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Peribunyaviridae; unclassified Peribunyaviridae
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
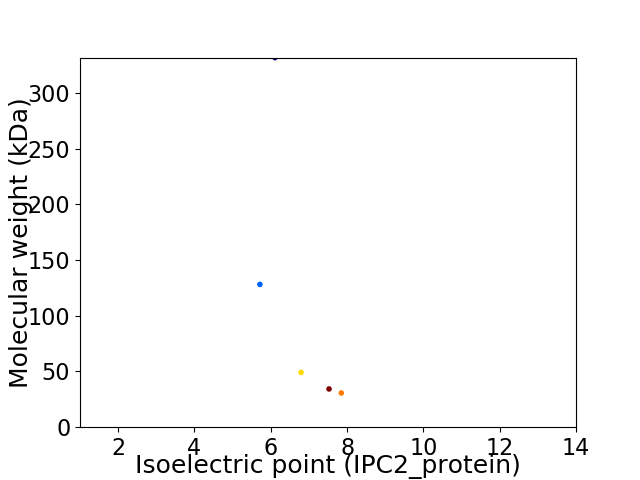
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5H8S9|A0A0C5H8S9_9VIRU NSs OS=Mulberry vein banding virus OX=1266451 GN=NSs PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 10.1KK3 pKa = 10.38YY4 pKa = 10.43YY5 pKa = 10.4LPSYY9 pKa = 10.57CLGLVLLFLVSEE21 pKa = 4.82VYY23 pKa = 10.75LINQMEE29 pKa = 4.77HH30 pKa = 6.7DD31 pKa = 4.13VQVKK35 pKa = 9.64RR36 pKa = 11.84FQSRR40 pKa = 11.84YY41 pKa = 9.7KK42 pKa = 10.84ADD44 pKa = 4.28DD45 pKa = 4.21PDD47 pKa = 4.95DD48 pKa = 3.86LVEE51 pKa = 4.99IEE53 pKa = 5.14EE54 pKa = 4.24PTEE57 pKa = 4.46SIVHH61 pKa = 5.02EE62 pKa = 4.16TRR64 pKa = 11.84FQKK67 pKa = 10.34KK68 pKa = 9.38KK69 pKa = 9.36KK70 pKa = 9.79LSRR73 pKa = 11.84ILRR76 pKa = 11.84DD77 pKa = 4.03DD78 pKa = 3.63EE79 pKa = 4.44ATTQSSASKK88 pKa = 10.31LDD90 pKa = 3.3CDD92 pKa = 3.78KK93 pKa = 11.35FEE95 pKa = 5.52KK96 pKa = 10.22RR97 pKa = 11.84YY98 pKa = 10.3CLIKK102 pKa = 10.68GVSDD106 pKa = 4.67FNAHH110 pKa = 5.16YY111 pKa = 10.54QLDD114 pKa = 3.85NGEE117 pKa = 4.69EE118 pKa = 4.11IISCISNSASIFEE131 pKa = 4.18IYY133 pKa = 10.16QYY135 pKa = 11.12EE136 pKa = 4.28KK137 pKa = 10.36EE138 pKa = 4.09FKK140 pKa = 10.25KK141 pKa = 10.84IKK143 pKa = 10.5FKK145 pKa = 11.29DD146 pKa = 3.72FLVVPVLKK154 pKa = 10.98LEE156 pKa = 4.19NKK158 pKa = 9.6KK159 pKa = 10.37VLEE162 pKa = 4.28VGTKK166 pKa = 9.75FFFVDD171 pKa = 3.86KK172 pKa = 11.02SNNPVNIDD180 pKa = 3.23PKK182 pKa = 11.28VNLKK186 pKa = 10.65SPTVSKK192 pKa = 10.95LSVRR196 pKa = 11.84LSGDD200 pKa = 3.35CKK202 pKa = 10.31INQVSMSTPYY212 pKa = 10.12QIKK215 pKa = 10.18LRR217 pKa = 11.84SEE219 pKa = 4.07EE220 pKa = 4.14NIGVLVKK227 pKa = 10.53NVKK230 pKa = 9.88DD231 pKa = 3.74PKK233 pKa = 10.28SGNVKK238 pKa = 9.78TIFGDD243 pKa = 3.53STINFQPEE251 pKa = 4.21EE252 pKa = 3.9LDD254 pKa = 3.63GNHH257 pKa = 6.69FLLCGDD263 pKa = 3.78KK264 pKa = 10.89SSLIMKK270 pKa = 9.73VDD272 pKa = 3.41VAVRR276 pKa = 11.84NCVSKK281 pKa = 11.45YY282 pKa = 9.76SDD284 pKa = 3.57EE285 pKa = 4.08PKK287 pKa = 10.64KK288 pKa = 10.85IFFCTNFSYY297 pKa = 10.01FKK299 pKa = 9.94WIFVFLMVAFPISWLVWKK317 pKa = 8.41TKK319 pKa = 9.99NALSIWYY326 pKa = 8.68DD327 pKa = 2.89IVGIVTYY334 pKa = 9.66PILWILNWFWPYY346 pKa = 10.73FPFKK350 pKa = 10.72CRR352 pKa = 11.84ICGCISFLTHH362 pKa = 6.29TCTEE366 pKa = 4.1KK367 pKa = 10.85CVCNQSEE374 pKa = 4.06ASKK377 pKa = 11.09DD378 pKa = 3.68HH379 pKa = 6.31TSEE382 pKa = 4.64CYY384 pKa = 10.31LFSRR388 pKa = 11.84DD389 pKa = 3.45KK390 pKa = 11.21KK391 pKa = 10.74DD392 pKa = 3.35WNEE395 pKa = 3.79LTLLQQFQFTINTKK409 pKa = 9.64ISTNFLVFVTKK420 pKa = 10.06MIIASILISYY430 pKa = 9.28IPSSIALQQSNLCVEE445 pKa = 4.06KK446 pKa = 10.54CYY448 pKa = 11.35YY449 pKa = 10.34NLNLDD454 pKa = 3.69SLTTDD459 pKa = 3.39RR460 pKa = 11.84FGLSKK465 pKa = 11.33NNFEE469 pKa = 4.33TCEE472 pKa = 3.98CSIGNVITEE481 pKa = 4.44TIYY484 pKa = 11.04RR485 pKa = 11.84EE486 pKa = 4.45GIPVSRR492 pKa = 11.84ATSLNDD498 pKa = 4.04CIVGSDD504 pKa = 3.24TCMVSDD510 pKa = 4.39NQAQNLFACRR520 pKa = 11.84NGCNSLNSIKK530 pKa = 10.59NIPDD534 pKa = 3.3TKK536 pKa = 10.39FSKK539 pKa = 10.47FYY541 pKa = 9.96KK542 pKa = 10.08GKK544 pKa = 10.02SFRR547 pKa = 11.84GNLTSLKK554 pKa = 9.48IANRR558 pKa = 11.84LRR560 pKa = 11.84EE561 pKa = 4.28GYY563 pKa = 8.76MDD565 pKa = 4.73SPTEE569 pKa = 4.08SKK571 pKa = 10.44ILEE574 pKa = 4.26EE575 pKa = 4.12EE576 pKa = 3.94STRR579 pKa = 11.84EE580 pKa = 3.72YY581 pKa = 11.24KK582 pKa = 10.42FYY584 pKa = 11.36SSLKK588 pKa = 10.0VDD590 pKa = 5.69DD591 pKa = 4.95IPPEE595 pKa = 3.94NLMPRR600 pKa = 11.84QSLVFSTEE608 pKa = 3.11VDD610 pKa = 3.09GKK612 pKa = 10.74YY613 pKa = 9.97RR614 pKa = 11.84YY615 pKa = 9.71LLEE618 pKa = 4.11MDD620 pKa = 4.09IKK622 pKa = 11.06ASTGSVYY629 pKa = 10.98LLNDD633 pKa = 4.31DD634 pKa = 4.88ASHH637 pKa = 7.11SPMEE641 pKa = 3.71FMVYY645 pKa = 9.25VKK647 pKa = 10.53SVGVEE652 pKa = 3.2YY653 pKa = 10.4DD654 pKa = 2.72IRR656 pKa = 11.84YY657 pKa = 9.56KK658 pKa = 11.11YY659 pKa = 9.59STAKK663 pKa = 10.07IDD665 pKa = 3.86TTVSDD670 pKa = 4.23YY671 pKa = 11.57LVTCTGKK678 pKa = 10.63CADD681 pKa = 5.18CIKK684 pKa = 10.37QKK686 pKa = 10.68PKK688 pKa = 10.93VGVLDD693 pKa = 4.06FCVTPTSWWGCEE705 pKa = 3.82EE706 pKa = 4.53LGCLAINEE714 pKa = 4.33GAICGHH720 pKa = 6.11CTNIYY725 pKa = 10.33DD726 pKa = 4.66LSSLINIYY734 pKa = 8.6QVIEE738 pKa = 4.08SHH740 pKa = 5.38VTAEE744 pKa = 4.13ICVKK748 pKa = 10.77SLDD751 pKa = 3.93GYY753 pKa = 9.1SCKK756 pKa = 10.14KK757 pKa = 10.17HH758 pKa = 6.52SDD760 pKa = 3.26RR761 pKa = 11.84SPIQTDD767 pKa = 3.94HH768 pKa = 6.5YY769 pKa = 9.97QLDD772 pKa = 3.87MSIDD776 pKa = 3.46LHH778 pKa = 7.51NDD780 pKa = 2.92YY781 pKa = 10.78MSTDD785 pKa = 3.08KK786 pKa = 11.27LFAVNKK792 pKa = 6.17QQKK795 pKa = 9.51VLTGNIADD803 pKa = 4.67LGDD806 pKa = 3.62FAGSSFGHH814 pKa = 5.78PQITVDD820 pKa = 4.2GVPLAVPAALTQNDD834 pKa = 4.24FSWSCSAVGEE844 pKa = 4.14KK845 pKa = 10.15KK846 pKa = 10.59INIRR850 pKa = 11.84QCGLYY855 pKa = 8.68TYY857 pKa = 11.35SMVYY861 pKa = 10.1VLSPSKK867 pKa = 10.88DD868 pKa = 3.33NSHH871 pKa = 6.49LQEE874 pKa = 5.2DD875 pKa = 4.13KK876 pKa = 11.39NKK878 pKa = 10.92LYY880 pKa = 9.99MEE882 pKa = 4.78KK883 pKa = 10.73DD884 pKa = 3.46FLVGKK889 pKa = 9.98LKK891 pKa = 10.25MVIDD895 pKa = 3.96MPKK898 pKa = 10.62EE899 pKa = 3.83MFKK902 pKa = 10.59KK903 pKa = 10.72VPTKK907 pKa = 10.34PVLSEE912 pKa = 3.96TKK914 pKa = 7.94MTCSGCTQCAVGIDD928 pKa = 3.83CNITYY933 pKa = 9.74TSDD936 pKa = 3.04TTFSSRR942 pKa = 11.84IMMDD946 pKa = 2.76SCSFKK951 pKa = 10.68SDD953 pKa = 3.31QLGTFLGPNEE963 pKa = 4.17KK964 pKa = 9.49TIKK967 pKa = 10.0AYY969 pKa = 10.5CSEE972 pKa = 4.54SIVDD976 pKa = 3.9KK977 pKa = 10.63SLKK980 pKa = 10.17FIPEE984 pKa = 4.16DD985 pKa = 3.94QEE987 pKa = 4.81DD988 pKa = 4.31LTVDD992 pKa = 3.24IQVDD996 pKa = 3.66EE997 pKa = 4.77FVQVDD1002 pKa = 3.77QDD1004 pKa = 4.31TIIHH1008 pKa = 6.59FDD1010 pKa = 3.87DD1011 pKa = 5.37KK1012 pKa = 11.3SAHH1015 pKa = 6.98DD1016 pKa = 4.17EE1017 pKa = 4.3NKK1019 pKa = 9.65HH1020 pKa = 6.34HH1021 pKa = 7.3SDD1023 pKa = 3.33TSISSLWDD1031 pKa = 3.04WVKK1034 pKa = 11.29APFNWVASFFGSFFDD1049 pKa = 4.86LIRR1052 pKa = 11.84IILVVLAICVGIYY1065 pKa = 10.04ILNCIFKK1072 pKa = 10.54LSKK1075 pKa = 9.55TYY1077 pKa = 10.93YY1078 pKa = 8.39IDD1080 pKa = 3.39KK1081 pKa = 10.01RR1082 pKa = 11.84RR1083 pKa = 11.84QRR1085 pKa = 11.84MEE1087 pKa = 4.01DD1088 pKa = 3.04AVEE1091 pKa = 4.48SIEE1094 pKa = 5.12SSVLLTNYY1102 pKa = 9.12TGLDD1106 pKa = 3.16QTRR1109 pKa = 11.84KK1110 pKa = 9.82RR1111 pKa = 11.84KK1112 pKa = 9.84SPPKK1116 pKa = 9.65GYY1118 pKa = 10.27EE1119 pKa = 3.6FSLEE1123 pKa = 4.06VV1124 pKa = 3.32
MM1 pKa = 7.49KK2 pKa = 10.1KK3 pKa = 10.38YY4 pKa = 10.43YY5 pKa = 10.4LPSYY9 pKa = 10.57CLGLVLLFLVSEE21 pKa = 4.82VYY23 pKa = 10.75LINQMEE29 pKa = 4.77HH30 pKa = 6.7DD31 pKa = 4.13VQVKK35 pKa = 9.64RR36 pKa = 11.84FQSRR40 pKa = 11.84YY41 pKa = 9.7KK42 pKa = 10.84ADD44 pKa = 4.28DD45 pKa = 4.21PDD47 pKa = 4.95DD48 pKa = 3.86LVEE51 pKa = 4.99IEE53 pKa = 5.14EE54 pKa = 4.24PTEE57 pKa = 4.46SIVHH61 pKa = 5.02EE62 pKa = 4.16TRR64 pKa = 11.84FQKK67 pKa = 10.34KK68 pKa = 9.38KK69 pKa = 9.36KK70 pKa = 9.79LSRR73 pKa = 11.84ILRR76 pKa = 11.84DD77 pKa = 4.03DD78 pKa = 3.63EE79 pKa = 4.44ATTQSSASKK88 pKa = 10.31LDD90 pKa = 3.3CDD92 pKa = 3.78KK93 pKa = 11.35FEE95 pKa = 5.52KK96 pKa = 10.22RR97 pKa = 11.84YY98 pKa = 10.3CLIKK102 pKa = 10.68GVSDD106 pKa = 4.67FNAHH110 pKa = 5.16YY111 pKa = 10.54QLDD114 pKa = 3.85NGEE117 pKa = 4.69EE118 pKa = 4.11IISCISNSASIFEE131 pKa = 4.18IYY133 pKa = 10.16QYY135 pKa = 11.12EE136 pKa = 4.28KK137 pKa = 10.36EE138 pKa = 4.09FKK140 pKa = 10.25KK141 pKa = 10.84IKK143 pKa = 10.5FKK145 pKa = 11.29DD146 pKa = 3.72FLVVPVLKK154 pKa = 10.98LEE156 pKa = 4.19NKK158 pKa = 9.6KK159 pKa = 10.37VLEE162 pKa = 4.28VGTKK166 pKa = 9.75FFFVDD171 pKa = 3.86KK172 pKa = 11.02SNNPVNIDD180 pKa = 3.23PKK182 pKa = 11.28VNLKK186 pKa = 10.65SPTVSKK192 pKa = 10.95LSVRR196 pKa = 11.84LSGDD200 pKa = 3.35CKK202 pKa = 10.31INQVSMSTPYY212 pKa = 10.12QIKK215 pKa = 10.18LRR217 pKa = 11.84SEE219 pKa = 4.07EE220 pKa = 4.14NIGVLVKK227 pKa = 10.53NVKK230 pKa = 9.88DD231 pKa = 3.74PKK233 pKa = 10.28SGNVKK238 pKa = 9.78TIFGDD243 pKa = 3.53STINFQPEE251 pKa = 4.21EE252 pKa = 3.9LDD254 pKa = 3.63GNHH257 pKa = 6.69FLLCGDD263 pKa = 3.78KK264 pKa = 10.89SSLIMKK270 pKa = 9.73VDD272 pKa = 3.41VAVRR276 pKa = 11.84NCVSKK281 pKa = 11.45YY282 pKa = 9.76SDD284 pKa = 3.57EE285 pKa = 4.08PKK287 pKa = 10.64KK288 pKa = 10.85IFFCTNFSYY297 pKa = 10.01FKK299 pKa = 9.94WIFVFLMVAFPISWLVWKK317 pKa = 8.41TKK319 pKa = 9.99NALSIWYY326 pKa = 8.68DD327 pKa = 2.89IVGIVTYY334 pKa = 9.66PILWILNWFWPYY346 pKa = 10.73FPFKK350 pKa = 10.72CRR352 pKa = 11.84ICGCISFLTHH362 pKa = 6.29TCTEE366 pKa = 4.1KK367 pKa = 10.85CVCNQSEE374 pKa = 4.06ASKK377 pKa = 11.09DD378 pKa = 3.68HH379 pKa = 6.31TSEE382 pKa = 4.64CYY384 pKa = 10.31LFSRR388 pKa = 11.84DD389 pKa = 3.45KK390 pKa = 11.21KK391 pKa = 10.74DD392 pKa = 3.35WNEE395 pKa = 3.79LTLLQQFQFTINTKK409 pKa = 9.64ISTNFLVFVTKK420 pKa = 10.06MIIASILISYY430 pKa = 9.28IPSSIALQQSNLCVEE445 pKa = 4.06KK446 pKa = 10.54CYY448 pKa = 11.35YY449 pKa = 10.34NLNLDD454 pKa = 3.69SLTTDD459 pKa = 3.39RR460 pKa = 11.84FGLSKK465 pKa = 11.33NNFEE469 pKa = 4.33TCEE472 pKa = 3.98CSIGNVITEE481 pKa = 4.44TIYY484 pKa = 11.04RR485 pKa = 11.84EE486 pKa = 4.45GIPVSRR492 pKa = 11.84ATSLNDD498 pKa = 4.04CIVGSDD504 pKa = 3.24TCMVSDD510 pKa = 4.39NQAQNLFACRR520 pKa = 11.84NGCNSLNSIKK530 pKa = 10.59NIPDD534 pKa = 3.3TKK536 pKa = 10.39FSKK539 pKa = 10.47FYY541 pKa = 9.96KK542 pKa = 10.08GKK544 pKa = 10.02SFRR547 pKa = 11.84GNLTSLKK554 pKa = 9.48IANRR558 pKa = 11.84LRR560 pKa = 11.84EE561 pKa = 4.28GYY563 pKa = 8.76MDD565 pKa = 4.73SPTEE569 pKa = 4.08SKK571 pKa = 10.44ILEE574 pKa = 4.26EE575 pKa = 4.12EE576 pKa = 3.94STRR579 pKa = 11.84EE580 pKa = 3.72YY581 pKa = 11.24KK582 pKa = 10.42FYY584 pKa = 11.36SSLKK588 pKa = 10.0VDD590 pKa = 5.69DD591 pKa = 4.95IPPEE595 pKa = 3.94NLMPRR600 pKa = 11.84QSLVFSTEE608 pKa = 3.11VDD610 pKa = 3.09GKK612 pKa = 10.74YY613 pKa = 9.97RR614 pKa = 11.84YY615 pKa = 9.71LLEE618 pKa = 4.11MDD620 pKa = 4.09IKK622 pKa = 11.06ASTGSVYY629 pKa = 10.98LLNDD633 pKa = 4.31DD634 pKa = 4.88ASHH637 pKa = 7.11SPMEE641 pKa = 3.71FMVYY645 pKa = 9.25VKK647 pKa = 10.53SVGVEE652 pKa = 3.2YY653 pKa = 10.4DD654 pKa = 2.72IRR656 pKa = 11.84YY657 pKa = 9.56KK658 pKa = 11.11YY659 pKa = 9.59STAKK663 pKa = 10.07IDD665 pKa = 3.86TTVSDD670 pKa = 4.23YY671 pKa = 11.57LVTCTGKK678 pKa = 10.63CADD681 pKa = 5.18CIKK684 pKa = 10.37QKK686 pKa = 10.68PKK688 pKa = 10.93VGVLDD693 pKa = 4.06FCVTPTSWWGCEE705 pKa = 3.82EE706 pKa = 4.53LGCLAINEE714 pKa = 4.33GAICGHH720 pKa = 6.11CTNIYY725 pKa = 10.33DD726 pKa = 4.66LSSLINIYY734 pKa = 8.6QVIEE738 pKa = 4.08SHH740 pKa = 5.38VTAEE744 pKa = 4.13ICVKK748 pKa = 10.77SLDD751 pKa = 3.93GYY753 pKa = 9.1SCKK756 pKa = 10.14KK757 pKa = 10.17HH758 pKa = 6.52SDD760 pKa = 3.26RR761 pKa = 11.84SPIQTDD767 pKa = 3.94HH768 pKa = 6.5YY769 pKa = 9.97QLDD772 pKa = 3.87MSIDD776 pKa = 3.46LHH778 pKa = 7.51NDD780 pKa = 2.92YY781 pKa = 10.78MSTDD785 pKa = 3.08KK786 pKa = 11.27LFAVNKK792 pKa = 6.17QQKK795 pKa = 9.51VLTGNIADD803 pKa = 4.67LGDD806 pKa = 3.62FAGSSFGHH814 pKa = 5.78PQITVDD820 pKa = 4.2GVPLAVPAALTQNDD834 pKa = 4.24FSWSCSAVGEE844 pKa = 4.14KK845 pKa = 10.15KK846 pKa = 10.59INIRR850 pKa = 11.84QCGLYY855 pKa = 8.68TYY857 pKa = 11.35SMVYY861 pKa = 10.1VLSPSKK867 pKa = 10.88DD868 pKa = 3.33NSHH871 pKa = 6.49LQEE874 pKa = 5.2DD875 pKa = 4.13KK876 pKa = 11.39NKK878 pKa = 10.92LYY880 pKa = 9.99MEE882 pKa = 4.78KK883 pKa = 10.73DD884 pKa = 3.46FLVGKK889 pKa = 9.98LKK891 pKa = 10.25MVIDD895 pKa = 3.96MPKK898 pKa = 10.62EE899 pKa = 3.83MFKK902 pKa = 10.59KK903 pKa = 10.72VPTKK907 pKa = 10.34PVLSEE912 pKa = 3.96TKK914 pKa = 7.94MTCSGCTQCAVGIDD928 pKa = 3.83CNITYY933 pKa = 9.74TSDD936 pKa = 3.04TTFSSRR942 pKa = 11.84IMMDD946 pKa = 2.76SCSFKK951 pKa = 10.68SDD953 pKa = 3.31QLGTFLGPNEE963 pKa = 4.17KK964 pKa = 9.49TIKK967 pKa = 10.0AYY969 pKa = 10.5CSEE972 pKa = 4.54SIVDD976 pKa = 3.9KK977 pKa = 10.63SLKK980 pKa = 10.17FIPEE984 pKa = 4.16DD985 pKa = 3.94QEE987 pKa = 4.81DD988 pKa = 4.31LTVDD992 pKa = 3.24IQVDD996 pKa = 3.66EE997 pKa = 4.77FVQVDD1002 pKa = 3.77QDD1004 pKa = 4.31TIIHH1008 pKa = 6.59FDD1010 pKa = 3.87DD1011 pKa = 5.37KK1012 pKa = 11.3SAHH1015 pKa = 6.98DD1016 pKa = 4.17EE1017 pKa = 4.3NKK1019 pKa = 9.65HH1020 pKa = 6.34HH1021 pKa = 7.3SDD1023 pKa = 3.33TSISSLWDD1031 pKa = 3.04WVKK1034 pKa = 11.29APFNWVASFFGSFFDD1049 pKa = 4.86LIRR1052 pKa = 11.84IILVVLAICVGIYY1065 pKa = 10.04ILNCIFKK1072 pKa = 10.54LSKK1075 pKa = 9.55TYY1077 pKa = 10.93YY1078 pKa = 8.39IDD1080 pKa = 3.39KK1081 pKa = 10.01RR1082 pKa = 11.84RR1083 pKa = 11.84QRR1085 pKa = 11.84MEE1087 pKa = 4.01DD1088 pKa = 3.04AVEE1091 pKa = 4.48SIEE1094 pKa = 5.12SSVLLTNYY1102 pKa = 9.12TGLDD1106 pKa = 3.16QTRR1109 pKa = 11.84KK1110 pKa = 9.82RR1111 pKa = 11.84KK1112 pKa = 9.84SPPKK1116 pKa = 9.65GYY1118 pKa = 10.27EE1119 pKa = 3.6FSLEE1123 pKa = 4.06VV1124 pKa = 3.32
Molecular weight: 128.14 kDa
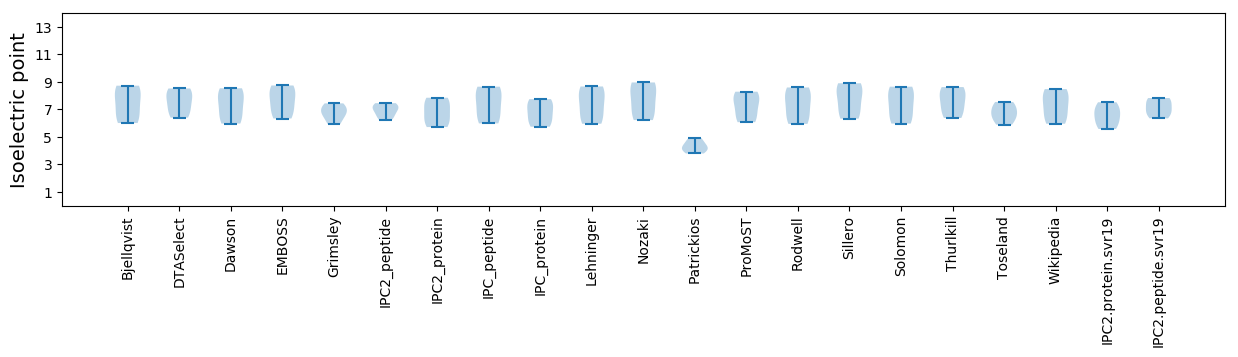
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9UTA1|K9UTA1_9VIRU Nucleoprotein OS=Mulberry vein banding virus OX=1266451 GN=N PE=3 SV=1
MM1 pKa = 6.79STVRR5 pKa = 11.84QLSEE9 pKa = 4.2SKK11 pKa = 8.9ITALLAGGKK20 pKa = 9.91ADD22 pKa = 3.89VEE24 pKa = 4.42IEE26 pKa = 4.37TEE28 pKa = 3.97EE29 pKa = 4.13STPGFNFKK37 pKa = 11.0AFFDD41 pKa = 4.0KK42 pKa = 11.31NHH44 pKa = 6.51DD45 pKa = 3.77VEE47 pKa = 4.81ITFATCLNILKK58 pKa = 10.14SRR60 pKa = 11.84KK61 pKa = 9.54QIFAACKK68 pKa = 7.51NGKK71 pKa = 9.75YY72 pKa = 10.09NFCGHH77 pKa = 6.77NIIATTANAGIDD89 pKa = 3.23DD90 pKa = 3.94WTFKK94 pKa = 9.99RR95 pKa = 11.84TEE97 pKa = 3.48ALIRR101 pKa = 11.84VKK103 pKa = 9.93MIRR106 pKa = 11.84MVEE109 pKa = 3.92KK110 pKa = 10.42AASNEE115 pKa = 4.25SKK117 pKa = 10.56LAMYY121 pKa = 9.46QKK123 pKa = 10.8VMEE126 pKa = 4.83LPLVSAYY133 pKa = 10.41GLNAPPEE140 pKa = 4.26FNACALRR147 pKa = 11.84LMLCIGGPLSLLSSFKK163 pKa = 10.84ALAPIVFPLAYY174 pKa = 8.49YY175 pKa = 10.63QNVKK179 pKa = 10.31KK180 pKa = 10.66EE181 pKa = 3.8KK182 pKa = 10.6LGIKK186 pKa = 10.06NFSTYY191 pKa = 9.83EE192 pKa = 3.92QVCKK196 pKa = 9.96VAKK199 pKa = 9.93VLSAAQFDD207 pKa = 4.1FQGDD211 pKa = 4.14FKK213 pKa = 10.94TLFEE217 pKa = 4.59EE218 pKa = 4.34AVKK221 pKa = 9.97MLSEE225 pKa = 4.61SNPGTASSISLKK237 pKa = 10.73KK238 pKa = 10.13YY239 pKa = 10.29DD240 pKa = 3.9EE241 pKa = 4.12QVKK244 pKa = 10.67YY245 pKa = 8.93MDD247 pKa = 4.24KK248 pKa = 10.69VFSSSLVVDD257 pKa = 4.91DD258 pKa = 4.72YY259 pKa = 12.27GEE261 pKa = 4.01NSKK264 pKa = 10.83KK265 pKa = 10.26KK266 pKa = 9.67SQPSTSNSIEE276 pKa = 3.93VV277 pKa = 3.25
MM1 pKa = 6.79STVRR5 pKa = 11.84QLSEE9 pKa = 4.2SKK11 pKa = 8.9ITALLAGGKK20 pKa = 9.91ADD22 pKa = 3.89VEE24 pKa = 4.42IEE26 pKa = 4.37TEE28 pKa = 3.97EE29 pKa = 4.13STPGFNFKK37 pKa = 11.0AFFDD41 pKa = 4.0KK42 pKa = 11.31NHH44 pKa = 6.51DD45 pKa = 3.77VEE47 pKa = 4.81ITFATCLNILKK58 pKa = 10.14SRR60 pKa = 11.84KK61 pKa = 9.54QIFAACKK68 pKa = 7.51NGKK71 pKa = 9.75YY72 pKa = 10.09NFCGHH77 pKa = 6.77NIIATTANAGIDD89 pKa = 3.23DD90 pKa = 3.94WTFKK94 pKa = 9.99RR95 pKa = 11.84TEE97 pKa = 3.48ALIRR101 pKa = 11.84VKK103 pKa = 9.93MIRR106 pKa = 11.84MVEE109 pKa = 3.92KK110 pKa = 10.42AASNEE115 pKa = 4.25SKK117 pKa = 10.56LAMYY121 pKa = 9.46QKK123 pKa = 10.8VMEE126 pKa = 4.83LPLVSAYY133 pKa = 10.41GLNAPPEE140 pKa = 4.26FNACALRR147 pKa = 11.84LMLCIGGPLSLLSSFKK163 pKa = 10.84ALAPIVFPLAYY174 pKa = 8.49YY175 pKa = 10.63QNVKK179 pKa = 10.31KK180 pKa = 10.66EE181 pKa = 3.8KK182 pKa = 10.6LGIKK186 pKa = 10.06NFSTYY191 pKa = 9.83EE192 pKa = 3.92QVCKK196 pKa = 9.96VAKK199 pKa = 9.93VLSAAQFDD207 pKa = 4.1FQGDD211 pKa = 4.14FKK213 pKa = 10.94TLFEE217 pKa = 4.59EE218 pKa = 4.34AVKK221 pKa = 9.97MLSEE225 pKa = 4.61SNPGTASSISLKK237 pKa = 10.73KK238 pKa = 10.13YY239 pKa = 10.29DD240 pKa = 3.9EE241 pKa = 4.12QVKK244 pKa = 10.67YY245 pKa = 8.93MDD247 pKa = 4.24KK248 pKa = 10.69VFSSSLVVDD257 pKa = 4.91DD258 pKa = 4.72YY259 pKa = 12.27GEE261 pKa = 4.01NSKK264 pKa = 10.83KK265 pKa = 10.26KK266 pKa = 9.67SQPSTSNSIEE276 pKa = 3.93VV277 pKa = 3.25
Molecular weight: 30.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5026 |
277 |
2877 |
1005.2 |
114.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.278 ± 1.055 | 2.189 ± 0.506 |
5.969 ± 0.417 | 6.526 ± 0.395 |
4.895 ± 0.287 | 4.019 ± 0.383 |
1.83 ± 0.26 | 7.461 ± 0.364 |
8.894 ± 0.256 | 9.272 ± 0.575 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.402 ± 0.585 | 6.327 ± 0.441 |
2.865 ± 0.284 | 2.945 ± 0.168 |
3.243 ± 0.299 | 9.471 ± 0.384 |
5.591 ± 0.158 | 5.949 ± 0.699 |
0.816 ± 0.174 | 4.059 ± 0.386 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
