
Pacific flying fox faeces associated circular DNA virus-2
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.03
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A140CTV3|A0A140CTV3_9VIRU Replication-associated protein OS=Pacific flying fox faeces associated circular DNA virus-2 OX=1796011 PE=3 SV=1
MM1 pKa = 7.86ASPPSRR7 pKa = 11.84QRR9 pKa = 11.84QARR12 pKa = 11.84NIIKK16 pKa = 10.18GLAVCRR22 pKa = 11.84VPPAPEE28 pKa = 3.72TSFQMVGPIRR38 pKa = 11.84QRR40 pKa = 11.84NFRR43 pKa = 11.84INATRR48 pKa = 11.84FLLTYY53 pKa = 8.65PQSTFDD59 pKa = 3.8VQLVWDD65 pKa = 5.11FFNSLAIKK73 pKa = 9.38PKK75 pKa = 10.1RR76 pKa = 11.84AILCRR81 pKa = 11.84EE82 pKa = 3.82HH83 pKa = 7.25HH84 pKa = 6.22QDD86 pKa = 3.04GSEE89 pKa = 4.13HH90 pKa = 4.38VHH92 pKa = 5.35VAIEE96 pKa = 3.84FEE98 pKa = 4.22RR99 pKa = 11.84AVNTTRR105 pKa = 11.84VSIFDD110 pKa = 3.49FGGRR114 pKa = 11.84HH115 pKa = 6.13PNIQSARR122 pKa = 11.84NWAACVNYY130 pKa = 9.52CRR132 pKa = 11.84KK133 pKa = 10.16EE134 pKa = 4.1GVIEE138 pKa = 3.82TAYY141 pKa = 10.45FGCTAEE147 pKa = 4.44DD148 pKa = 3.55ATVASAPGEE157 pKa = 4.04RR158 pKa = 11.84APAEE162 pKa = 4.17SPYY165 pKa = 11.2ARR167 pKa = 11.84AEE169 pKa = 3.9SARR172 pKa = 11.84TIRR175 pKa = 11.84EE176 pKa = 3.71WFEE179 pKa = 3.43YY180 pKa = 10.4CIAEE184 pKa = 4.8GIGYY188 pKa = 10.15AYY190 pKa = 10.54ANAIWNQIHH199 pKa = 5.82SVRR202 pKa = 11.84PPTYY206 pKa = 10.17FEE208 pKa = 4.77NDD210 pKa = 2.77FSGIVSGPTLSHH222 pKa = 6.6LRR224 pKa = 11.84WDD226 pKa = 4.6PEE228 pKa = 3.41WHH230 pKa = 6.71TIFLCGPSGLGKK242 pKa = 10.53SSWALANAPVPFLFVTDD259 pKa = 3.71IDD261 pKa = 4.11DD262 pKa = 5.1LGFFDD267 pKa = 5.39PEE269 pKa = 3.87VHH271 pKa = 6.57KK272 pKa = 11.1ALVFDD277 pKa = 5.39EE278 pKa = 4.47IRR280 pKa = 11.84CTGEE284 pKa = 3.93PNDD287 pKa = 4.15NSAKK291 pKa = 10.43GKK293 pKa = 9.58WPLTSQIKK301 pKa = 9.82LATWDD306 pKa = 3.81TPVSIRR312 pKa = 11.84IRR314 pKa = 11.84YY315 pKa = 8.61KK316 pKa = 9.69IAHH319 pKa = 6.92LPAHH323 pKa = 5.71VPKK326 pKa = 10.59IFTSCDD332 pKa = 3.08WFPMNGDD339 pKa = 3.26EE340 pKa = 5.19QIRR343 pKa = 11.84RR344 pKa = 11.84RR345 pKa = 11.84IKK347 pKa = 10.73AFNLYY352 pKa = 10.16EE353 pKa = 4.72DD354 pKa = 4.13GRR356 pKa = 11.84STEE359 pKa = 4.26SLWLL363 pKa = 3.9
MM1 pKa = 7.86ASPPSRR7 pKa = 11.84QRR9 pKa = 11.84QARR12 pKa = 11.84NIIKK16 pKa = 10.18GLAVCRR22 pKa = 11.84VPPAPEE28 pKa = 3.72TSFQMVGPIRR38 pKa = 11.84QRR40 pKa = 11.84NFRR43 pKa = 11.84INATRR48 pKa = 11.84FLLTYY53 pKa = 8.65PQSTFDD59 pKa = 3.8VQLVWDD65 pKa = 5.11FFNSLAIKK73 pKa = 9.38PKK75 pKa = 10.1RR76 pKa = 11.84AILCRR81 pKa = 11.84EE82 pKa = 3.82HH83 pKa = 7.25HH84 pKa = 6.22QDD86 pKa = 3.04GSEE89 pKa = 4.13HH90 pKa = 4.38VHH92 pKa = 5.35VAIEE96 pKa = 3.84FEE98 pKa = 4.22RR99 pKa = 11.84AVNTTRR105 pKa = 11.84VSIFDD110 pKa = 3.49FGGRR114 pKa = 11.84HH115 pKa = 6.13PNIQSARR122 pKa = 11.84NWAACVNYY130 pKa = 9.52CRR132 pKa = 11.84KK133 pKa = 10.16EE134 pKa = 4.1GVIEE138 pKa = 3.82TAYY141 pKa = 10.45FGCTAEE147 pKa = 4.44DD148 pKa = 3.55ATVASAPGEE157 pKa = 4.04RR158 pKa = 11.84APAEE162 pKa = 4.17SPYY165 pKa = 11.2ARR167 pKa = 11.84AEE169 pKa = 3.9SARR172 pKa = 11.84TIRR175 pKa = 11.84EE176 pKa = 3.71WFEE179 pKa = 3.43YY180 pKa = 10.4CIAEE184 pKa = 4.8GIGYY188 pKa = 10.15AYY190 pKa = 10.54ANAIWNQIHH199 pKa = 5.82SVRR202 pKa = 11.84PPTYY206 pKa = 10.17FEE208 pKa = 4.77NDD210 pKa = 2.77FSGIVSGPTLSHH222 pKa = 6.6LRR224 pKa = 11.84WDD226 pKa = 4.6PEE228 pKa = 3.41WHH230 pKa = 6.71TIFLCGPSGLGKK242 pKa = 10.53SSWALANAPVPFLFVTDD259 pKa = 3.71IDD261 pKa = 4.11DD262 pKa = 5.1LGFFDD267 pKa = 5.39PEE269 pKa = 3.87VHH271 pKa = 6.57KK272 pKa = 11.1ALVFDD277 pKa = 5.39EE278 pKa = 4.47IRR280 pKa = 11.84CTGEE284 pKa = 3.93PNDD287 pKa = 4.15NSAKK291 pKa = 10.43GKK293 pKa = 9.58WPLTSQIKK301 pKa = 9.82LATWDD306 pKa = 3.81TPVSIRR312 pKa = 11.84IRR314 pKa = 11.84YY315 pKa = 8.61KK316 pKa = 9.69IAHH319 pKa = 6.92LPAHH323 pKa = 5.71VPKK326 pKa = 10.59IFTSCDD332 pKa = 3.08WFPMNGDD339 pKa = 3.26EE340 pKa = 5.19QIRR343 pKa = 11.84RR344 pKa = 11.84RR345 pKa = 11.84IKK347 pKa = 10.73AFNLYY352 pKa = 10.16EE353 pKa = 4.72DD354 pKa = 4.13GRR356 pKa = 11.84STEE359 pKa = 4.26SLWLL363 pKa = 3.9
Molecular weight: 41.16 kDa
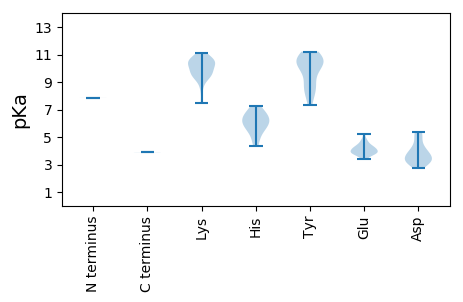
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTV3|A0A140CTV3_9VIRU Replication-associated protein OS=Pacific flying fox faeces associated circular DNA virus-2 OX=1796011 PE=3 SV=1
MM1 pKa = 7.86PGQLQRR7 pKa = 11.84YY8 pKa = 8.31RR9 pKa = 11.84RR10 pKa = 11.84GSLALGSAYY19 pKa = 10.52RR20 pKa = 11.84LAKK23 pKa = 10.52AGWKK27 pKa = 9.61AGRR30 pKa = 11.84YY31 pKa = 7.78LAQNINRR38 pKa = 11.84GRR40 pKa = 11.84SMSTLRR46 pKa = 11.84RR47 pKa = 11.84SSGTGGTSRR56 pKa = 11.84RR57 pKa = 11.84GTKK60 pKa = 9.4RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84TSSGGRR69 pKa = 11.84RR70 pKa = 11.84GSYY73 pKa = 10.74QMGSMKK79 pKa = 10.39KK80 pKa = 8.46MRR82 pKa = 11.84GGFSGSRR89 pKa = 11.84DD90 pKa = 3.46GVSSFSTKK98 pKa = 9.56RR99 pKa = 11.84KK100 pKa = 9.79RR101 pKa = 11.84GTKK104 pKa = 10.02RR105 pKa = 11.84GWKK108 pKa = 9.45KK109 pKa = 10.39VLAPQVLRR117 pKa = 11.84TVTSGRR123 pKa = 11.84ILRR126 pKa = 11.84NDD128 pKa = 3.26SGLQTVHH135 pKa = 6.08EE136 pKa = 4.47VLMTNAIPNAVNTTALMSGTTDD158 pKa = 4.56LEE160 pKa = 3.79RR161 pKa = 11.84LYY163 pKa = 11.05QLLRR167 pKa = 11.84VKK169 pKa = 10.68DD170 pKa = 3.92PNTEE174 pKa = 3.86FGQAANATITRR185 pKa = 11.84RR186 pKa = 11.84FFVDD190 pKa = 3.16SCRR193 pKa = 11.84VKK195 pKa = 9.82ITFKK199 pKa = 10.48NQTSIPVEE207 pKa = 3.77LTLYY211 pKa = 10.72DD212 pKa = 3.17ITSRR216 pKa = 11.84KK217 pKa = 10.3DD218 pKa = 3.1NTSALPRR225 pKa = 11.84YY226 pKa = 7.3TPAGSWLEE234 pKa = 3.87GLTNEE239 pKa = 4.39SATFSSEE246 pKa = 4.02PNDD249 pKa = 3.23NAQYY253 pKa = 9.77STFPGSKK260 pKa = 9.18PFEE263 pKa = 4.18SVKK266 pKa = 10.15FCQLFNVRR274 pKa = 11.84KK275 pKa = 8.93SKK277 pKa = 10.71HH278 pKa = 4.92MLLGAGEE285 pKa = 4.15THH287 pKa = 5.62IHH289 pKa = 6.26YY290 pKa = 10.67INLNFKK296 pKa = 10.81YY297 pKa = 9.46MFNRR301 pKa = 11.84EE302 pKa = 4.12EE303 pKa = 3.88FSGFYY308 pKa = 8.8QLRR311 pKa = 11.84NVTQWCLAVLKK322 pKa = 10.65GGIVSSGAAITGPAQVSSSYY342 pKa = 11.21GIVDD346 pKa = 3.58YY347 pKa = 8.41TTEE350 pKa = 3.57AVYY353 pKa = 10.51KK354 pKa = 7.48FKK356 pKa = 11.06CMEE359 pKa = 4.18KK360 pKa = 9.32STTVYY365 pKa = 10.68QSFNALPAIAPAQQTSILEE384 pKa = 4.38DD385 pKa = 3.33TDD387 pKa = 3.73LRR389 pKa = 11.84AVLQTAA395 pKa = 3.89
MM1 pKa = 7.86PGQLQRR7 pKa = 11.84YY8 pKa = 8.31RR9 pKa = 11.84RR10 pKa = 11.84GSLALGSAYY19 pKa = 10.52RR20 pKa = 11.84LAKK23 pKa = 10.52AGWKK27 pKa = 9.61AGRR30 pKa = 11.84YY31 pKa = 7.78LAQNINRR38 pKa = 11.84GRR40 pKa = 11.84SMSTLRR46 pKa = 11.84RR47 pKa = 11.84SSGTGGTSRR56 pKa = 11.84RR57 pKa = 11.84GTKK60 pKa = 9.4RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84TSSGGRR69 pKa = 11.84RR70 pKa = 11.84GSYY73 pKa = 10.74QMGSMKK79 pKa = 10.39KK80 pKa = 8.46MRR82 pKa = 11.84GGFSGSRR89 pKa = 11.84DD90 pKa = 3.46GVSSFSTKK98 pKa = 9.56RR99 pKa = 11.84KK100 pKa = 9.79RR101 pKa = 11.84GTKK104 pKa = 10.02RR105 pKa = 11.84GWKK108 pKa = 9.45KK109 pKa = 10.39VLAPQVLRR117 pKa = 11.84TVTSGRR123 pKa = 11.84ILRR126 pKa = 11.84NDD128 pKa = 3.26SGLQTVHH135 pKa = 6.08EE136 pKa = 4.47VLMTNAIPNAVNTTALMSGTTDD158 pKa = 4.56LEE160 pKa = 3.79RR161 pKa = 11.84LYY163 pKa = 11.05QLLRR167 pKa = 11.84VKK169 pKa = 10.68DD170 pKa = 3.92PNTEE174 pKa = 3.86FGQAANATITRR185 pKa = 11.84RR186 pKa = 11.84FFVDD190 pKa = 3.16SCRR193 pKa = 11.84VKK195 pKa = 9.82ITFKK199 pKa = 10.48NQTSIPVEE207 pKa = 3.77LTLYY211 pKa = 10.72DD212 pKa = 3.17ITSRR216 pKa = 11.84KK217 pKa = 10.3DD218 pKa = 3.1NTSALPRR225 pKa = 11.84YY226 pKa = 7.3TPAGSWLEE234 pKa = 3.87GLTNEE239 pKa = 4.39SATFSSEE246 pKa = 4.02PNDD249 pKa = 3.23NAQYY253 pKa = 9.77STFPGSKK260 pKa = 9.18PFEE263 pKa = 4.18SVKK266 pKa = 10.15FCQLFNVRR274 pKa = 11.84KK275 pKa = 8.93SKK277 pKa = 10.71HH278 pKa = 4.92MLLGAGEE285 pKa = 4.15THH287 pKa = 5.62IHH289 pKa = 6.26YY290 pKa = 10.67INLNFKK296 pKa = 10.81YY297 pKa = 9.46MFNRR301 pKa = 11.84EE302 pKa = 4.12EE303 pKa = 3.88FSGFYY308 pKa = 8.8QLRR311 pKa = 11.84NVTQWCLAVLKK322 pKa = 10.65GGIVSSGAAITGPAQVSSSYY342 pKa = 11.21GIVDD346 pKa = 3.58YY347 pKa = 8.41TTEE350 pKa = 3.57AVYY353 pKa = 10.51KK354 pKa = 7.48FKK356 pKa = 11.06CMEE359 pKa = 4.18KK360 pKa = 9.32STTVYY365 pKa = 10.68QSFNALPAIAPAQQTSILEE384 pKa = 4.38DD385 pKa = 3.33TDD387 pKa = 3.73LRR389 pKa = 11.84AVLQTAA395 pKa = 3.89
Molecular weight: 43.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
758 |
363 |
395 |
379.0 |
42.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.443 ± 0.869 | 1.715 ± 0.554 |
3.694 ± 0.717 | 4.749 ± 0.95 |
5.145 ± 0.664 | 7.256 ± 1.266 |
1.979 ± 0.762 | 5.277 ± 1.367 |
4.617 ± 0.951 | 6.86 ± 0.979 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.715 ± 0.644 | 4.617 ± 0.152 |
5.145 ± 1.462 | 3.826 ± 0.577 |
8.311 ± 0.433 | 8.443 ± 1.328 |
7.52 ± 1.457 | 5.409 ± 0.073 |
1.979 ± 0.762 | 3.298 ± 0.394 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
