
Human papillomavirus type 137
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 16
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
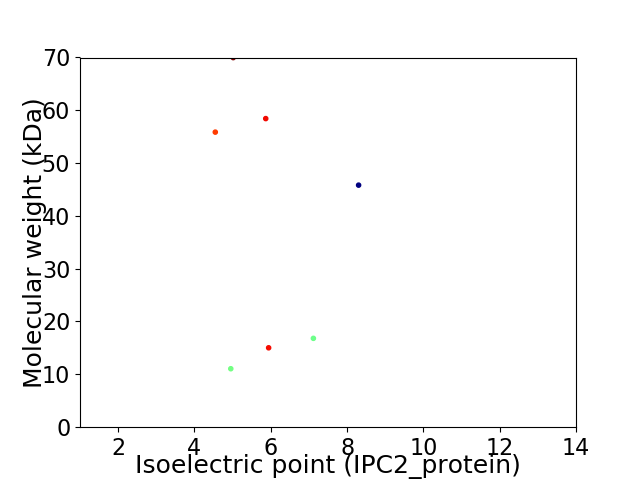
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3P6M3|I3P6M3_9PAPI Major capsid protein L1 OS=Human papillomavirus type 137 OX=1070410 GN=L1 PE=3 SV=1
MM1 pKa = 7.07QANKK5 pKa = 9.57RR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 8.32RR9 pKa = 11.84AAVEE13 pKa = 4.12DD14 pKa = 4.67IYY16 pKa = 11.66AKK18 pKa = 10.82GCTQPGGYY26 pKa = 9.78CPPDD30 pKa = 3.29VKK32 pKa = 11.21NKK34 pKa = 10.1VEE36 pKa = 4.16GNTWADD42 pKa = 3.41FLLKK46 pKa = 10.8VFGSVVYY53 pKa = 10.04FGGLGIGTGKK63 pKa = 8.78GTGGSTGYY71 pKa = 8.7TPLGGTVGSRR81 pKa = 11.84GTTNTIKK88 pKa = 9.55PTIPLDD94 pKa = 3.65PLGVPDD100 pKa = 5.02IVTVDD105 pKa = 4.64PIAPEE110 pKa = 3.85AASIVPLAEE119 pKa = 4.24GLPEE123 pKa = 4.8PGVIDD128 pKa = 3.71TGTSFPGLAADD139 pKa = 4.09NEE141 pKa = 4.68NIVTVLDD148 pKa = 3.99PLSEE152 pKa = 4.1VTGVGEE158 pKa = 4.44HH159 pKa = 6.8PNIITGGTADD169 pKa = 4.07SPAILDD175 pKa = 4.04VQTSPPPAKK184 pKa = 10.25KK185 pKa = 9.96ILLDD189 pKa = 3.62PSISKK194 pKa = 7.22TTTAVQTHH202 pKa = 6.94ASHH205 pKa = 7.25VDD207 pKa = 2.93ANLNIFVDD215 pKa = 3.91AQSFGTHH222 pKa = 4.75VGYY225 pKa = 9.07TEE227 pKa = 5.89DD228 pKa = 4.59IPLEE232 pKa = 4.37EE233 pKa = 4.25INLRR237 pKa = 11.84SEE239 pKa = 4.62FEE241 pKa = 4.61LEE243 pKa = 3.89DD244 pKa = 4.11SEE246 pKa = 5.25PKK248 pKa = 10.22TSTPFAEE255 pKa = 4.02RR256 pKa = 11.84VLNKK260 pKa = 9.38TKK262 pKa = 10.41QLYY265 pKa = 9.64SKK267 pKa = 10.34YY268 pKa = 8.56VQQVPTRR275 pKa = 11.84PAEE278 pKa = 3.91FALYY282 pKa = 9.08TSRR285 pKa = 11.84FEE287 pKa = 4.4FEE289 pKa = 4.0NPAFEE294 pKa = 4.82EE295 pKa = 4.3DD296 pKa = 3.26VTMEE300 pKa = 4.34FEE302 pKa = 4.21NDD304 pKa = 3.1LAEE307 pKa = 4.96IGEE310 pKa = 4.36ITTPAVSDD318 pKa = 3.51VRR320 pKa = 11.84ILNRR324 pKa = 11.84PIYY327 pKa = 10.62SEE329 pKa = 3.91TADD332 pKa = 3.45RR333 pKa = 11.84TVRR336 pKa = 11.84ISRR339 pKa = 11.84LGQRR343 pKa = 11.84AGMKK347 pKa = 8.71TRR349 pKa = 11.84SGLEE353 pKa = 3.11IGQRR357 pKa = 11.84VHH359 pKa = 7.42FYY361 pKa = 10.99FDD363 pKa = 3.54LSDD366 pKa = 3.56IPRR369 pKa = 11.84EE370 pKa = 4.16SIEE373 pKa = 3.95LNTYY377 pKa = 10.18GNYY380 pKa = 9.35SHH382 pKa = 7.41EE383 pKa = 4.27STIVDD388 pKa = 3.76EE389 pKa = 4.81LLSSTFINPFEE400 pKa = 4.29MPVDD404 pKa = 3.79SEE406 pKa = 4.06IFAEE410 pKa = 4.95NEE412 pKa = 3.7LLDD415 pKa = 4.24PLEE418 pKa = 4.01EE419 pKa = 4.5DD420 pKa = 4.14FRR422 pKa = 11.84DD423 pKa = 3.54SHH425 pKa = 7.84IVVPYY430 pKa = 11.03LEE432 pKa = 5.47DD433 pKa = 3.41EE434 pKa = 4.67QINITPTLPPGLGLKK449 pKa = 10.04VYY451 pKa = 10.74SDD453 pKa = 3.54LSEE456 pKa = 4.77RR457 pKa = 11.84DD458 pKa = 3.62LLIHH462 pKa = 6.37YY463 pKa = 7.43PVQHH467 pKa = 7.02ADD469 pKa = 2.71IMVPDD474 pKa = 4.26TPYY477 pKa = 10.71IPVQPPDD484 pKa = 3.5GVLVDD489 pKa = 4.6DD490 pKa = 4.41NDD492 pKa = 4.27YY493 pKa = 10.95YY494 pKa = 11.45LHH496 pKa = 7.24PGLYY500 pKa = 8.83SRR502 pKa = 11.84KK503 pKa = 8.98RR504 pKa = 11.84KK505 pKa = 9.64RR506 pKa = 11.84RR507 pKa = 11.84VLL509 pKa = 3.52
MM1 pKa = 7.07QANKK5 pKa = 9.57RR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 8.32RR9 pKa = 11.84AAVEE13 pKa = 4.12DD14 pKa = 4.67IYY16 pKa = 11.66AKK18 pKa = 10.82GCTQPGGYY26 pKa = 9.78CPPDD30 pKa = 3.29VKK32 pKa = 11.21NKK34 pKa = 10.1VEE36 pKa = 4.16GNTWADD42 pKa = 3.41FLLKK46 pKa = 10.8VFGSVVYY53 pKa = 10.04FGGLGIGTGKK63 pKa = 8.78GTGGSTGYY71 pKa = 8.7TPLGGTVGSRR81 pKa = 11.84GTTNTIKK88 pKa = 9.55PTIPLDD94 pKa = 3.65PLGVPDD100 pKa = 5.02IVTVDD105 pKa = 4.64PIAPEE110 pKa = 3.85AASIVPLAEE119 pKa = 4.24GLPEE123 pKa = 4.8PGVIDD128 pKa = 3.71TGTSFPGLAADD139 pKa = 4.09NEE141 pKa = 4.68NIVTVLDD148 pKa = 3.99PLSEE152 pKa = 4.1VTGVGEE158 pKa = 4.44HH159 pKa = 6.8PNIITGGTADD169 pKa = 4.07SPAILDD175 pKa = 4.04VQTSPPPAKK184 pKa = 10.25KK185 pKa = 9.96ILLDD189 pKa = 3.62PSISKK194 pKa = 7.22TTTAVQTHH202 pKa = 6.94ASHH205 pKa = 7.25VDD207 pKa = 2.93ANLNIFVDD215 pKa = 3.91AQSFGTHH222 pKa = 4.75VGYY225 pKa = 9.07TEE227 pKa = 5.89DD228 pKa = 4.59IPLEE232 pKa = 4.37EE233 pKa = 4.25INLRR237 pKa = 11.84SEE239 pKa = 4.62FEE241 pKa = 4.61LEE243 pKa = 3.89DD244 pKa = 4.11SEE246 pKa = 5.25PKK248 pKa = 10.22TSTPFAEE255 pKa = 4.02RR256 pKa = 11.84VLNKK260 pKa = 9.38TKK262 pKa = 10.41QLYY265 pKa = 9.64SKK267 pKa = 10.34YY268 pKa = 8.56VQQVPTRR275 pKa = 11.84PAEE278 pKa = 3.91FALYY282 pKa = 9.08TSRR285 pKa = 11.84FEE287 pKa = 4.4FEE289 pKa = 4.0NPAFEE294 pKa = 4.82EE295 pKa = 4.3DD296 pKa = 3.26VTMEE300 pKa = 4.34FEE302 pKa = 4.21NDD304 pKa = 3.1LAEE307 pKa = 4.96IGEE310 pKa = 4.36ITTPAVSDD318 pKa = 3.51VRR320 pKa = 11.84ILNRR324 pKa = 11.84PIYY327 pKa = 10.62SEE329 pKa = 3.91TADD332 pKa = 3.45RR333 pKa = 11.84TVRR336 pKa = 11.84ISRR339 pKa = 11.84LGQRR343 pKa = 11.84AGMKK347 pKa = 8.71TRR349 pKa = 11.84SGLEE353 pKa = 3.11IGQRR357 pKa = 11.84VHH359 pKa = 7.42FYY361 pKa = 10.99FDD363 pKa = 3.54LSDD366 pKa = 3.56IPRR369 pKa = 11.84EE370 pKa = 4.16SIEE373 pKa = 3.95LNTYY377 pKa = 10.18GNYY380 pKa = 9.35SHH382 pKa = 7.41EE383 pKa = 4.27STIVDD388 pKa = 3.76EE389 pKa = 4.81LLSSTFINPFEE400 pKa = 4.29MPVDD404 pKa = 3.79SEE406 pKa = 4.06IFAEE410 pKa = 4.95NEE412 pKa = 3.7LLDD415 pKa = 4.24PLEE418 pKa = 4.01EE419 pKa = 4.5DD420 pKa = 4.14FRR422 pKa = 11.84DD423 pKa = 3.54SHH425 pKa = 7.84IVVPYY430 pKa = 11.03LEE432 pKa = 5.47DD433 pKa = 3.41EE434 pKa = 4.67QINITPTLPPGLGLKK449 pKa = 10.04VYY451 pKa = 10.74SDD453 pKa = 3.54LSEE456 pKa = 4.77RR457 pKa = 11.84DD458 pKa = 3.62LLIHH462 pKa = 6.37YY463 pKa = 7.43PVQHH467 pKa = 7.02ADD469 pKa = 2.71IMVPDD474 pKa = 4.26TPYY477 pKa = 10.71IPVQPPDD484 pKa = 3.5GVLVDD489 pKa = 4.6DD490 pKa = 4.41NDD492 pKa = 4.27YY493 pKa = 10.95YY494 pKa = 11.45LHH496 pKa = 7.24PGLYY500 pKa = 8.83SRR502 pKa = 11.84KK503 pKa = 8.98RR504 pKa = 11.84KK505 pKa = 9.64RR506 pKa = 11.84RR507 pKa = 11.84VLL509 pKa = 3.52
Molecular weight: 55.82 kDa
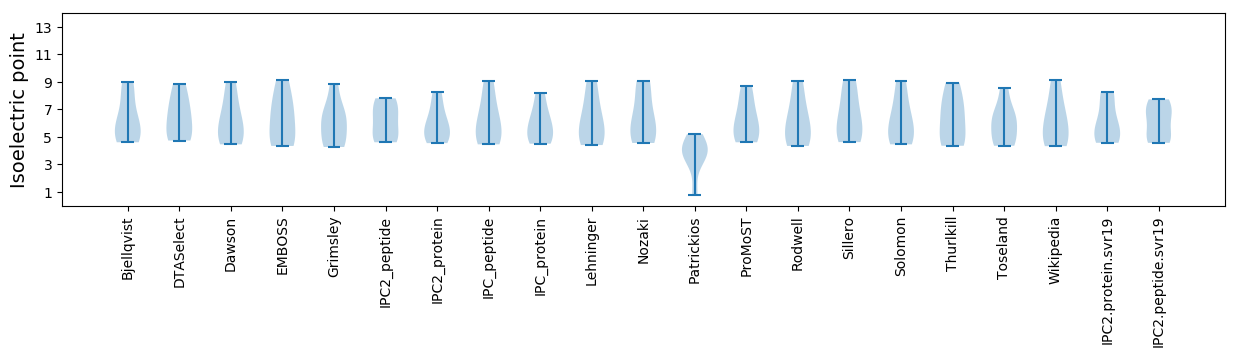
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3P6M1|I3P6M1_9PAPI Uncharacterized protein OS=Human papillomavirus type 137 OX=1070410 GN=E4 PE=4 SV=1
MM1 pKa = 7.14NQTDD5 pKa = 3.99LKK7 pKa = 10.99NRR9 pKa = 11.84LDD11 pKa = 4.04ALQDD15 pKa = 3.59ALMTLYY21 pKa = 10.76EE22 pKa = 4.91KK23 pKa = 11.1DD24 pKa = 3.76PVDD27 pKa = 4.08LSSQIEE33 pKa = 4.61HH34 pKa = 6.43YY35 pKa = 10.88SLLRR39 pKa = 11.84KK40 pKa = 9.21EE41 pKa = 5.61AVLQYY46 pKa = 8.89YY47 pKa = 9.14CRR49 pKa = 11.84KK50 pKa = 9.37EE51 pKa = 4.24GYY53 pKa = 9.2RR54 pKa = 11.84QLGLQILPSLTVSEE68 pKa = 5.01HH69 pKa = 5.28NAKK72 pKa = 10.09EE73 pKa = 4.08AIKK76 pKa = 9.89MVIILKK82 pKa = 10.28SLSKK86 pKa = 10.75SQFAGEE92 pKa = 3.92RR93 pKa = 11.84WTLRR97 pKa = 11.84DD98 pKa = 3.24TSLEE102 pKa = 4.1LFNTAPKK109 pKa = 10.81NCFKK113 pKa = 10.92KK114 pKa = 10.44DD115 pKa = 3.37GYY117 pKa = 10.78EE118 pKa = 3.83VLVWFDD124 pKa = 3.78HH125 pKa = 7.13DD126 pKa = 4.51PNNAYY131 pKa = 9.61PYY133 pKa = 9.04TNWNNIYY140 pKa = 10.07YY141 pKa = 10.46QGDD144 pKa = 3.25NDD146 pKa = 3.51VWYY149 pKa = 8.4KK150 pKa = 9.53TAGKK154 pKa = 10.32VDD156 pKa = 4.44LNGLYY161 pKa = 10.49YY162 pKa = 10.5EE163 pKa = 5.75DD164 pKa = 3.98YY165 pKa = 11.3DD166 pKa = 3.77NVKK169 pKa = 10.35VYY171 pKa = 9.89FVLFQEE177 pKa = 5.08KK178 pKa = 10.5ADD180 pKa = 4.37LYY182 pKa = 10.42SKK184 pKa = 10.53SGEE187 pKa = 3.73WTVNFKK193 pKa = 10.96NEE195 pKa = 4.03QLSSSIASSTRR206 pKa = 11.84RR207 pKa = 11.84PIPLLSHH214 pKa = 5.21EE215 pKa = 4.52TNRR218 pKa = 11.84NGNGASRR225 pKa = 11.84ATTRR229 pKa = 11.84NASPTSTALPISSYY243 pKa = 10.76GGRR246 pKa = 11.84EE247 pKa = 3.68EE248 pKa = 4.62SEE250 pKa = 4.42SEE252 pKa = 4.14EE253 pKa = 4.72GPSSTSRR260 pKa = 11.84SPTGRR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84GGEE273 pKa = 3.71QGEE276 pKa = 4.66SAARR280 pKa = 11.84PQKK283 pKa = 10.34RR284 pKa = 11.84KK285 pKa = 10.0RR286 pKa = 11.84GGDD289 pKa = 3.59AVPTAAEE296 pKa = 4.02VGSNHH301 pKa = 5.46RR302 pKa = 11.84TVPSTGLSRR311 pKa = 11.84LTRR314 pKa = 11.84LQAEE318 pKa = 4.16ARR320 pKa = 11.84DD321 pKa = 3.83PQLILINGAANQLKK335 pKa = 9.47CFRR338 pKa = 11.84YY339 pKa = 9.46RR340 pKa = 11.84CYY342 pKa = 11.2AKK344 pKa = 10.57AKK346 pKa = 9.83QNFIAMSTVWHH357 pKa = 6.09WVNNEE362 pKa = 3.84HH363 pKa = 7.1KK364 pKa = 9.89EE365 pKa = 4.22SSSKK369 pKa = 10.96LLVAFDD375 pKa = 3.95SDD377 pKa = 3.49KK378 pKa = 11.14QRR380 pKa = 11.84DD381 pKa = 3.91LFLVTTTLPKK391 pKa = 10.56GCTYY395 pKa = 10.9CFGQLDD401 pKa = 4.23CFF403 pKa = 4.57
MM1 pKa = 7.14NQTDD5 pKa = 3.99LKK7 pKa = 10.99NRR9 pKa = 11.84LDD11 pKa = 4.04ALQDD15 pKa = 3.59ALMTLYY21 pKa = 10.76EE22 pKa = 4.91KK23 pKa = 11.1DD24 pKa = 3.76PVDD27 pKa = 4.08LSSQIEE33 pKa = 4.61HH34 pKa = 6.43YY35 pKa = 10.88SLLRR39 pKa = 11.84KK40 pKa = 9.21EE41 pKa = 5.61AVLQYY46 pKa = 8.89YY47 pKa = 9.14CRR49 pKa = 11.84KK50 pKa = 9.37EE51 pKa = 4.24GYY53 pKa = 9.2RR54 pKa = 11.84QLGLQILPSLTVSEE68 pKa = 5.01HH69 pKa = 5.28NAKK72 pKa = 10.09EE73 pKa = 4.08AIKK76 pKa = 9.89MVIILKK82 pKa = 10.28SLSKK86 pKa = 10.75SQFAGEE92 pKa = 3.92RR93 pKa = 11.84WTLRR97 pKa = 11.84DD98 pKa = 3.24TSLEE102 pKa = 4.1LFNTAPKK109 pKa = 10.81NCFKK113 pKa = 10.92KK114 pKa = 10.44DD115 pKa = 3.37GYY117 pKa = 10.78EE118 pKa = 3.83VLVWFDD124 pKa = 3.78HH125 pKa = 7.13DD126 pKa = 4.51PNNAYY131 pKa = 9.61PYY133 pKa = 9.04TNWNNIYY140 pKa = 10.07YY141 pKa = 10.46QGDD144 pKa = 3.25NDD146 pKa = 3.51VWYY149 pKa = 8.4KK150 pKa = 9.53TAGKK154 pKa = 10.32VDD156 pKa = 4.44LNGLYY161 pKa = 10.49YY162 pKa = 10.5EE163 pKa = 5.75DD164 pKa = 3.98YY165 pKa = 11.3DD166 pKa = 3.77NVKK169 pKa = 10.35VYY171 pKa = 9.89FVLFQEE177 pKa = 5.08KK178 pKa = 10.5ADD180 pKa = 4.37LYY182 pKa = 10.42SKK184 pKa = 10.53SGEE187 pKa = 3.73WTVNFKK193 pKa = 10.96NEE195 pKa = 4.03QLSSSIASSTRR206 pKa = 11.84RR207 pKa = 11.84PIPLLSHH214 pKa = 5.21EE215 pKa = 4.52TNRR218 pKa = 11.84NGNGASRR225 pKa = 11.84ATTRR229 pKa = 11.84NASPTSTALPISSYY243 pKa = 10.76GGRR246 pKa = 11.84EE247 pKa = 3.68EE248 pKa = 4.62SEE250 pKa = 4.42SEE252 pKa = 4.14EE253 pKa = 4.72GPSSTSRR260 pKa = 11.84SPTGRR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84GGEE273 pKa = 3.71QGEE276 pKa = 4.66SAARR280 pKa = 11.84PQKK283 pKa = 10.34RR284 pKa = 11.84KK285 pKa = 10.0RR286 pKa = 11.84GGDD289 pKa = 3.59AVPTAAEE296 pKa = 4.02VGSNHH301 pKa = 5.46RR302 pKa = 11.84TVPSTGLSRR311 pKa = 11.84LTRR314 pKa = 11.84LQAEE318 pKa = 4.16ARR320 pKa = 11.84DD321 pKa = 3.83PQLILINGAANQLKK335 pKa = 9.47CFRR338 pKa = 11.84YY339 pKa = 9.46RR340 pKa = 11.84CYY342 pKa = 11.2AKK344 pKa = 10.57AKK346 pKa = 9.83QNFIAMSTVWHH357 pKa = 6.09WVNNEE362 pKa = 3.84HH363 pKa = 7.1KK364 pKa = 9.89EE365 pKa = 4.22SSSKK369 pKa = 10.96LLVAFDD375 pKa = 3.95SDD377 pKa = 3.49KK378 pKa = 11.14QRR380 pKa = 11.84DD381 pKa = 3.91LFLVTTTLPKK391 pKa = 10.56GCTYY395 pKa = 10.9CFGQLDD401 pKa = 4.23CFF403 pKa = 4.57
Molecular weight: 45.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2411 |
99 |
610 |
344.4 |
38.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.558 ± 0.345 | 2.323 ± 0.857 |
6.678 ± 0.444 | 6.512 ± 0.756 |
4.562 ± 0.519 | 5.226 ± 0.867 |
2.074 ± 0.144 | 4.936 ± 0.541 |
5.516 ± 0.753 | 10.079 ± 0.564 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.535 ± 0.35 | 5.309 ± 0.441 |
5.973 ± 1.168 | 4.023 ± 0.347 |
5.392 ± 0.684 | 7.507 ± 0.546 |
6.387 ± 0.458 | 5.682 ± 0.573 |
1.12 ± 0.317 | 3.608 ± 0.467 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
