
Halomonas sp. YLB-10
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Halomonas; unclassified Halomonas
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
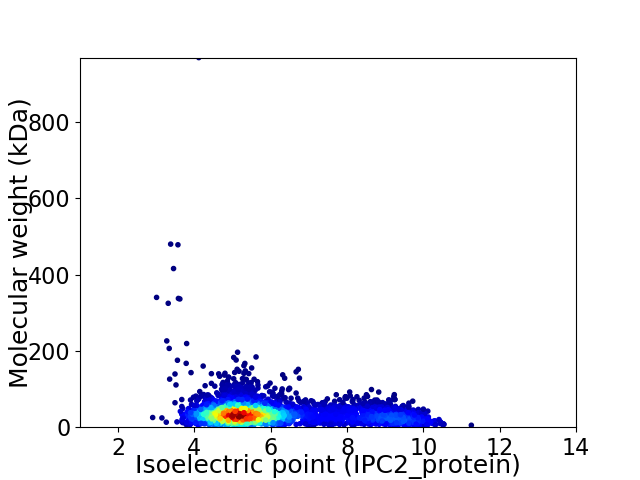
Virtual 2D-PAGE plot for 4189 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
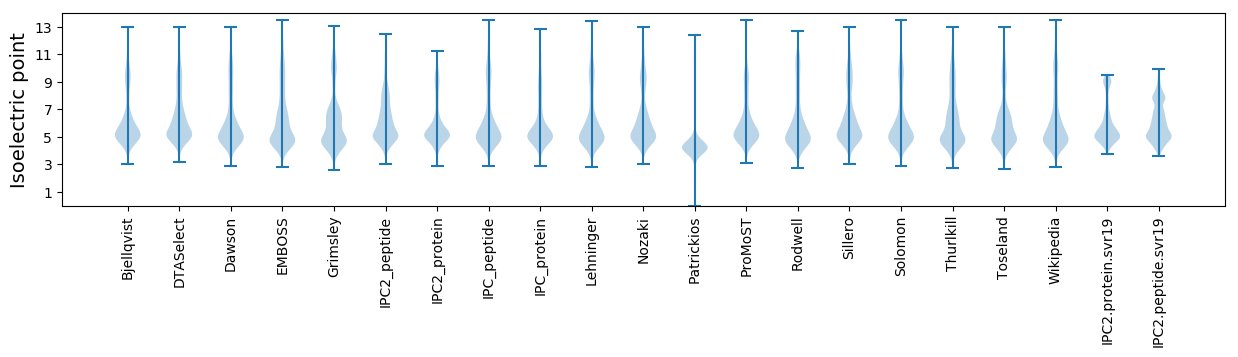
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N9U8K7|A0A3N9U8K7_9GAMM Sarcosine oxidase subunit alpha family protein OS=Halomonas sp. YLB-10 OX=2483111 GN=EBB56_02975 PE=3 SV=1
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGAAAFTAAGAQAATVYY27 pKa = 10.61NQDD30 pKa = 3.01GTKK33 pKa = 10.39LDD35 pKa = 3.23IYY37 pKa = 11.24GNVQIVYY44 pKa = 10.4AGVKK48 pKa = 10.55DD49 pKa = 3.93EE50 pKa = 5.49DD51 pKa = 3.55GHH53 pKa = 7.56SRR55 pKa = 11.84DD56 pKa = 4.17EE57 pKa = 4.16IADD60 pKa = 3.65NGSTFGFAAEE70 pKa = 3.86HH71 pKa = 6.08VIYY74 pKa = 10.81NGLIGYY80 pKa = 9.5AKK82 pKa = 10.18IEE84 pKa = 4.05IDD86 pKa = 4.24DD87 pKa = 4.44FKK89 pKa = 11.48ADD91 pKa = 3.47EE92 pKa = 4.32MKK94 pKa = 10.94VAGRR98 pKa = 11.84DD99 pKa = 3.57AGDD102 pKa = 3.16TAYY105 pKa = 9.75IGLKK109 pKa = 10.68GNFGDD114 pKa = 5.02ARR116 pKa = 11.84IGSYY120 pKa = 11.06DD121 pKa = 3.67SLLDD125 pKa = 4.29DD126 pKa = 5.74WIQDD130 pKa = 3.78PISNNEE136 pKa = 3.8YY137 pKa = 10.85FDD139 pKa = 4.53VSDD142 pKa = 3.85SNTFKK147 pKa = 10.97GAPGEE152 pKa = 4.43DD153 pKa = 3.68ADD155 pKa = 4.68DD156 pKa = 4.53ASVVAGDD163 pKa = 4.12PEE165 pKa = 4.71GDD167 pKa = 3.53KK168 pKa = 9.7ITYY171 pKa = 7.65TSPVWGGFQFALGTQYY187 pKa = 11.23KK188 pKa = 10.58GDD190 pKa = 3.99AEE192 pKa = 4.29NEE194 pKa = 3.94NLTDD198 pKa = 3.75GGEE201 pKa = 3.81ASIFGGLKK209 pKa = 8.16YY210 pKa = 9.34TIGGFSVAGVYY221 pKa = 10.53DD222 pKa = 3.76DD223 pKa = 5.34LGIYY227 pKa = 9.99DD228 pKa = 5.0IEE230 pKa = 4.52DD231 pKa = 3.19QSVSYY236 pKa = 11.02LSTDD240 pKa = 3.04VAADD244 pKa = 3.6GTVTTTQNNVTIDD257 pKa = 4.04DD258 pKa = 4.3EE259 pKa = 5.09DD260 pKa = 5.97LGDD263 pKa = 3.65QYY265 pKa = 11.56GVTMQYY271 pKa = 10.08QWDD274 pKa = 4.0SLRR277 pKa = 11.84VAVKK281 pKa = 9.05YY282 pKa = 10.67EE283 pKa = 3.95RR284 pKa = 11.84FEE286 pKa = 4.35SDD288 pKa = 3.28NDD290 pKa = 3.54FLKK293 pKa = 11.15DD294 pKa = 3.08ADD296 pKa = 4.07YY297 pKa = 10.92YY298 pKa = 11.51AIGARR303 pKa = 11.84YY304 pKa = 9.66GYY306 pKa = 10.32LNGMGDD312 pKa = 3.04IYY314 pKa = 11.2GSYY317 pKa = 10.48QYY319 pKa = 10.71IDD321 pKa = 3.53LGGSGFSDD329 pKa = 3.75FDD331 pKa = 4.12SANDD335 pKa = 5.25DD336 pKa = 4.5EE337 pKa = 6.45NDD339 pKa = 3.41DD340 pKa = 4.83DD341 pKa = 4.66NFNEE345 pKa = 4.15VVAGVTYY352 pKa = 10.62NISDD356 pKa = 3.12AMYY359 pKa = 8.48TYY361 pKa = 11.28VEE363 pKa = 4.17GAVRR367 pKa = 11.84DD368 pKa = 3.88RR369 pKa = 11.84DD370 pKa = 3.44NDD372 pKa = 3.44QGDD375 pKa = 4.49GYY377 pKa = 11.52AVGLTYY383 pKa = 11.05LFF385 pKa = 4.66
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGAAAFTAAGAQAATVYY27 pKa = 10.61NQDD30 pKa = 3.01GTKK33 pKa = 10.39LDD35 pKa = 3.23IYY37 pKa = 11.24GNVQIVYY44 pKa = 10.4AGVKK48 pKa = 10.55DD49 pKa = 3.93EE50 pKa = 5.49DD51 pKa = 3.55GHH53 pKa = 7.56SRR55 pKa = 11.84DD56 pKa = 4.17EE57 pKa = 4.16IADD60 pKa = 3.65NGSTFGFAAEE70 pKa = 3.86HH71 pKa = 6.08VIYY74 pKa = 10.81NGLIGYY80 pKa = 9.5AKK82 pKa = 10.18IEE84 pKa = 4.05IDD86 pKa = 4.24DD87 pKa = 4.44FKK89 pKa = 11.48ADD91 pKa = 3.47EE92 pKa = 4.32MKK94 pKa = 10.94VAGRR98 pKa = 11.84DD99 pKa = 3.57AGDD102 pKa = 3.16TAYY105 pKa = 9.75IGLKK109 pKa = 10.68GNFGDD114 pKa = 5.02ARR116 pKa = 11.84IGSYY120 pKa = 11.06DD121 pKa = 3.67SLLDD125 pKa = 4.29DD126 pKa = 5.74WIQDD130 pKa = 3.78PISNNEE136 pKa = 3.8YY137 pKa = 10.85FDD139 pKa = 4.53VSDD142 pKa = 3.85SNTFKK147 pKa = 10.97GAPGEE152 pKa = 4.43DD153 pKa = 3.68ADD155 pKa = 4.68DD156 pKa = 4.53ASVVAGDD163 pKa = 4.12PEE165 pKa = 4.71GDD167 pKa = 3.53KK168 pKa = 9.7ITYY171 pKa = 7.65TSPVWGGFQFALGTQYY187 pKa = 11.23KK188 pKa = 10.58GDD190 pKa = 3.99AEE192 pKa = 4.29NEE194 pKa = 3.94NLTDD198 pKa = 3.75GGEE201 pKa = 3.81ASIFGGLKK209 pKa = 8.16YY210 pKa = 9.34TIGGFSVAGVYY221 pKa = 10.53DD222 pKa = 3.76DD223 pKa = 5.34LGIYY227 pKa = 9.99DD228 pKa = 5.0IEE230 pKa = 4.52DD231 pKa = 3.19QSVSYY236 pKa = 11.02LSTDD240 pKa = 3.04VAADD244 pKa = 3.6GTVTTTQNNVTIDD257 pKa = 4.04DD258 pKa = 4.3EE259 pKa = 5.09DD260 pKa = 5.97LGDD263 pKa = 3.65QYY265 pKa = 11.56GVTMQYY271 pKa = 10.08QWDD274 pKa = 4.0SLRR277 pKa = 11.84VAVKK281 pKa = 9.05YY282 pKa = 10.67EE283 pKa = 3.95RR284 pKa = 11.84FEE286 pKa = 4.35SDD288 pKa = 3.28NDD290 pKa = 3.54FLKK293 pKa = 11.15DD294 pKa = 3.08ADD296 pKa = 4.07YY297 pKa = 10.92YY298 pKa = 11.51AIGARR303 pKa = 11.84YY304 pKa = 9.66GYY306 pKa = 10.32LNGMGDD312 pKa = 3.04IYY314 pKa = 11.2GSYY317 pKa = 10.48QYY319 pKa = 10.71IDD321 pKa = 3.53LGGSGFSDD329 pKa = 3.75FDD331 pKa = 4.12SANDD335 pKa = 5.25DD336 pKa = 4.5EE337 pKa = 6.45NDD339 pKa = 3.41DD340 pKa = 4.83DD341 pKa = 4.66NFNEE345 pKa = 4.15VVAGVTYY352 pKa = 10.62NISDD356 pKa = 3.12AMYY359 pKa = 8.48TYY361 pKa = 11.28VEE363 pKa = 4.17GAVRR367 pKa = 11.84DD368 pKa = 3.88RR369 pKa = 11.84DD370 pKa = 3.44NDD372 pKa = 3.44QGDD375 pKa = 4.49GYY377 pKa = 11.52AVGLTYY383 pKa = 11.05LFF385 pKa = 4.66
Molecular weight: 41.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N9U1D3|A0A3N9U1D3_9GAMM Transcriptional regulator OS=Halomonas sp. YLB-10 OX=2483111 GN=EBB56_13285 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1430958 |
28 |
9628 |
341.6 |
37.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.248 ± 0.047 | 0.951 ± 0.014 |
6.239 ± 0.066 | 6.303 ± 0.042 |
3.366 ± 0.027 | 8.482 ± 0.049 |
2.354 ± 0.022 | 4.524 ± 0.03 |
2.517 ± 0.029 | 11.371 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.415 ± 0.028 | 2.427 ± 0.025 |
4.893 ± 0.036 | 3.745 ± 0.025 |
7.128 ± 0.053 | 5.93 ± 0.043 |
5.037 ± 0.046 | 7.368 ± 0.033 |
1.44 ± 0.017 | 2.262 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
