
Isfahan virus (ISFV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Vesiculovirus; Isfahan vesiculovirus
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
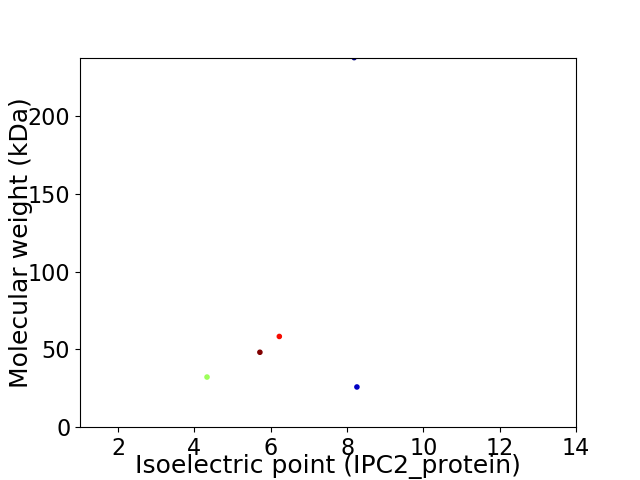
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q5K2K7|NCAP_ISFV Nucleoprotein OS=Isfahan virus OX=290008 GN=N PE=3 SV=1
MM1 pKa = 7.54SRR3 pKa = 11.84LNQILKK9 pKa = 10.27DD10 pKa = 4.0YY11 pKa = 9.9PLLEE15 pKa = 4.4ATTSEE20 pKa = 4.23IEE22 pKa = 4.27SMEE25 pKa = 4.18SSLADD30 pKa = 4.4DD31 pKa = 5.06VITSNDD37 pKa = 3.57DD38 pKa = 3.74EE39 pKa = 4.52IQSVSPQYY47 pKa = 10.84YY48 pKa = 9.87LRR50 pKa = 11.84DD51 pKa = 3.51MFKK54 pKa = 11.17ASITEE59 pKa = 4.2GPDD62 pKa = 3.27DD63 pKa = 5.26DD64 pKa = 4.97FPPVPEE70 pKa = 4.25VEE72 pKa = 3.95NDD74 pKa = 3.26IILDD78 pKa = 3.92DD79 pKa = 4.3DD80 pKa = 4.27EE81 pKa = 6.36EE82 pKa = 5.8YY83 pKa = 11.06DD84 pKa = 3.77GYY86 pKa = 11.53KK87 pKa = 10.27VDD89 pKa = 4.23FAEE92 pKa = 4.25ARR94 pKa = 11.84PWTALTQKK102 pKa = 10.84NIDD105 pKa = 2.88GRR107 pKa = 11.84MNLEE111 pKa = 3.91LMAPEE116 pKa = 4.72NLTDD120 pKa = 3.51AQYY123 pKa = 9.99KK124 pKa = 8.64QWVEE128 pKa = 4.28SVSSIMTISRR138 pKa = 11.84QIRR141 pKa = 11.84LHH143 pKa = 5.01QAEE146 pKa = 4.42IMDD149 pKa = 4.16TSSGLLIIEE158 pKa = 4.25NMIPSIGRR166 pKa = 11.84TSEE169 pKa = 3.93FKK171 pKa = 10.7SIPEE175 pKa = 4.83HH176 pKa = 6.47IPPSPTSDD184 pKa = 2.72HH185 pKa = 5.67TTPPSSLRR193 pKa = 11.84SDD195 pKa = 3.78TPSQTSSSSMGLPDD209 pKa = 4.12VSSASDD215 pKa = 3.2WSGMINKK222 pKa = 9.71KK223 pKa = 9.79IRR225 pKa = 11.84IPPVVSSKK233 pKa = 10.99SPYY236 pKa = 10.57EE237 pKa = 3.88FTLSDD242 pKa = 4.25LYY244 pKa = 11.46GSNQAALDD252 pKa = 3.95YY253 pKa = 11.11LSGSGMDD260 pKa = 4.76LRR262 pKa = 11.84TAVCSGLKK270 pKa = 9.48QRR272 pKa = 11.84GIYY275 pKa = 9.78NRR277 pKa = 11.84IRR279 pKa = 11.84IQYY282 pKa = 9.2KK283 pKa = 8.03ITPEE287 pKa = 4.03FVV289 pKa = 2.79
MM1 pKa = 7.54SRR3 pKa = 11.84LNQILKK9 pKa = 10.27DD10 pKa = 4.0YY11 pKa = 9.9PLLEE15 pKa = 4.4ATTSEE20 pKa = 4.23IEE22 pKa = 4.27SMEE25 pKa = 4.18SSLADD30 pKa = 4.4DD31 pKa = 5.06VITSNDD37 pKa = 3.57DD38 pKa = 3.74EE39 pKa = 4.52IQSVSPQYY47 pKa = 10.84YY48 pKa = 9.87LRR50 pKa = 11.84DD51 pKa = 3.51MFKK54 pKa = 11.17ASITEE59 pKa = 4.2GPDD62 pKa = 3.27DD63 pKa = 5.26DD64 pKa = 4.97FPPVPEE70 pKa = 4.25VEE72 pKa = 3.95NDD74 pKa = 3.26IILDD78 pKa = 3.92DD79 pKa = 4.3DD80 pKa = 4.27EE81 pKa = 6.36EE82 pKa = 5.8YY83 pKa = 11.06DD84 pKa = 3.77GYY86 pKa = 11.53KK87 pKa = 10.27VDD89 pKa = 4.23FAEE92 pKa = 4.25ARR94 pKa = 11.84PWTALTQKK102 pKa = 10.84NIDD105 pKa = 2.88GRR107 pKa = 11.84MNLEE111 pKa = 3.91LMAPEE116 pKa = 4.72NLTDD120 pKa = 3.51AQYY123 pKa = 9.99KK124 pKa = 8.64QWVEE128 pKa = 4.28SVSSIMTISRR138 pKa = 11.84QIRR141 pKa = 11.84LHH143 pKa = 5.01QAEE146 pKa = 4.42IMDD149 pKa = 4.16TSSGLLIIEE158 pKa = 4.25NMIPSIGRR166 pKa = 11.84TSEE169 pKa = 3.93FKK171 pKa = 10.7SIPEE175 pKa = 4.83HH176 pKa = 6.47IPPSPTSDD184 pKa = 2.72HH185 pKa = 5.67TTPPSSLRR193 pKa = 11.84SDD195 pKa = 3.78TPSQTSSSSMGLPDD209 pKa = 4.12VSSASDD215 pKa = 3.2WSGMINKK222 pKa = 9.71KK223 pKa = 9.79IRR225 pKa = 11.84IPPVVSSKK233 pKa = 10.99SPYY236 pKa = 10.57EE237 pKa = 3.88FTLSDD242 pKa = 4.25LYY244 pKa = 11.46GSNQAALDD252 pKa = 3.95YY253 pKa = 11.11LSGSGMDD260 pKa = 4.76LRR262 pKa = 11.84TAVCSGLKK270 pKa = 9.48QRR272 pKa = 11.84GIYY275 pKa = 9.78NRR277 pKa = 11.84IRR279 pKa = 11.84IQYY282 pKa = 9.2KK283 pKa = 8.03ITPEE287 pKa = 4.03FVV289 pKa = 2.79
Molecular weight: 32.25 kDa
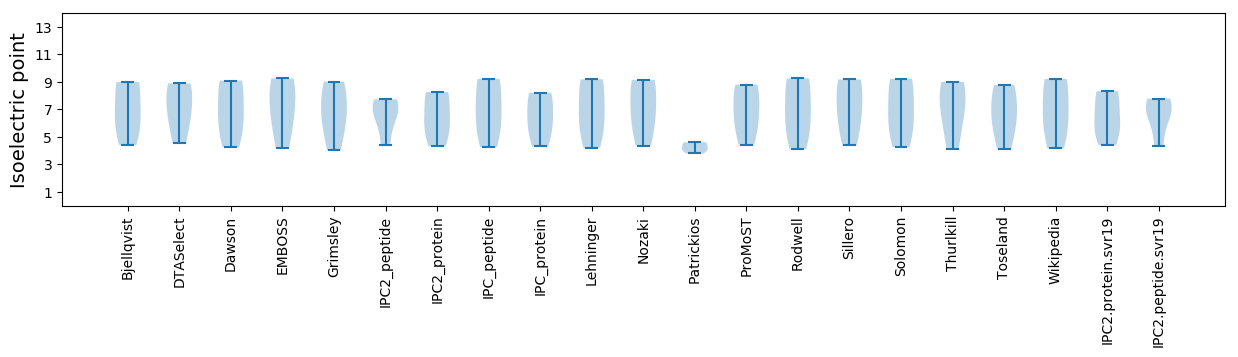
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q5K2K6|PHOSP_ISFV Phosphoprotein OS=Isfahan virus OX=290008 GN=P PE=3 SV=1
MM1 pKa = 7.65KK2 pKa = 10.29SLKK5 pKa = 10.36RR6 pKa = 11.84LIKK9 pKa = 10.25SNKK12 pKa = 9.18KK13 pKa = 10.4KK14 pKa = 10.56GDD16 pKa = 3.48KK17 pKa = 10.69KK18 pKa = 10.8NVSFMDD24 pKa = 3.08WDD26 pKa = 4.02EE27 pKa = 4.36PPSYY31 pKa = 10.83SDD33 pKa = 3.79SRR35 pKa = 11.84YY36 pKa = 9.61GCYY39 pKa = 9.75PSAPLFGVDD48 pKa = 4.99EE49 pKa = 4.31MMEE52 pKa = 4.2TLPTLGIQSLKK63 pKa = 10.18IQYY66 pKa = 9.51KK67 pKa = 10.24CSLQVRR73 pKa = 11.84AEE75 pKa = 4.25TPFTSFNDD83 pKa = 3.44AARR86 pKa = 11.84CISLWEE92 pKa = 3.83TDD94 pKa = 3.25YY95 pKa = 11.39RR96 pKa = 11.84GYY98 pKa = 11.0AGKK101 pKa = 10.57KK102 pKa = 7.81PFYY105 pKa = 9.88RR106 pKa = 11.84LLMLIATKK114 pKa = 10.44KK115 pKa = 10.22LRR117 pKa = 11.84AAPMSLMDD125 pKa = 4.22GNRR128 pKa = 11.84PEE130 pKa = 4.34YY131 pKa = 10.89SSMIQGQSIVHH142 pKa = 6.45HH143 pKa = 6.18SLGVIPPMMHH153 pKa = 5.84VPEE156 pKa = 4.47TFTRR160 pKa = 11.84EE161 pKa = 3.74WNLLTNKK168 pKa = 9.96GMITAQIWLGITDD181 pKa = 4.04VVDD184 pKa = 4.68DD185 pKa = 5.35LNPLINPALFSDD197 pKa = 4.33EE198 pKa = 5.29KK199 pKa = 11.35EE200 pKa = 4.07MTLTSQMFGLEE211 pKa = 3.99LKK213 pKa = 10.28KK214 pKa = 11.04RR215 pKa = 11.84NDD217 pKa = 3.59NTWLISKK224 pKa = 10.14SYY226 pKa = 10.85
MM1 pKa = 7.65KK2 pKa = 10.29SLKK5 pKa = 10.36RR6 pKa = 11.84LIKK9 pKa = 10.25SNKK12 pKa = 9.18KK13 pKa = 10.4KK14 pKa = 10.56GDD16 pKa = 3.48KK17 pKa = 10.69KK18 pKa = 10.8NVSFMDD24 pKa = 3.08WDD26 pKa = 4.02EE27 pKa = 4.36PPSYY31 pKa = 10.83SDD33 pKa = 3.79SRR35 pKa = 11.84YY36 pKa = 9.61GCYY39 pKa = 9.75PSAPLFGVDD48 pKa = 4.99EE49 pKa = 4.31MMEE52 pKa = 4.2TLPTLGIQSLKK63 pKa = 10.18IQYY66 pKa = 9.51KK67 pKa = 10.24CSLQVRR73 pKa = 11.84AEE75 pKa = 4.25TPFTSFNDD83 pKa = 3.44AARR86 pKa = 11.84CISLWEE92 pKa = 3.83TDD94 pKa = 3.25YY95 pKa = 11.39RR96 pKa = 11.84GYY98 pKa = 11.0AGKK101 pKa = 10.57KK102 pKa = 7.81PFYY105 pKa = 9.88RR106 pKa = 11.84LLMLIATKK114 pKa = 10.44KK115 pKa = 10.22LRR117 pKa = 11.84AAPMSLMDD125 pKa = 4.22GNRR128 pKa = 11.84PEE130 pKa = 4.34YY131 pKa = 10.89SSMIQGQSIVHH142 pKa = 6.45HH143 pKa = 6.18SLGVIPPMMHH153 pKa = 5.84VPEE156 pKa = 4.47TFTRR160 pKa = 11.84EE161 pKa = 3.74WNLLTNKK168 pKa = 9.96GMITAQIWLGITDD181 pKa = 4.04VVDD184 pKa = 4.68DD185 pKa = 5.35LNPLINPALFSDD197 pKa = 4.33EE198 pKa = 5.29KK199 pKa = 11.35EE200 pKa = 4.07MTLTSQMFGLEE211 pKa = 3.99LKK213 pKa = 10.28KK214 pKa = 11.04RR215 pKa = 11.84NDD217 pKa = 3.59NTWLISKK224 pKa = 10.14SYY226 pKa = 10.85
Molecular weight: 25.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3554 |
226 |
2093 |
710.8 |
80.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.952 ± 0.461 | 1.716 ± 0.288 |
5.937 ± 0.642 | 5.318 ± 0.327 |
3.995 ± 0.339 | 5.993 ± 0.394 |
2.279 ± 0.335 | 6.415 ± 0.554 |
6.528 ± 0.423 | 10.27 ± 0.626 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.926 ± 0.466 | 4.024 ± 0.335 |
4.53 ± 0.746 | 3.63 ± 0.276 |
5.121 ± 0.446 | 9.567 ± 0.765 |
5.656 ± 0.149 | 5.712 ± 0.696 |
1.97 ± 0.176 | 3.461 ± 0.214 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
