
Anatilimnocola aggregata
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae; Anatilimnocola
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
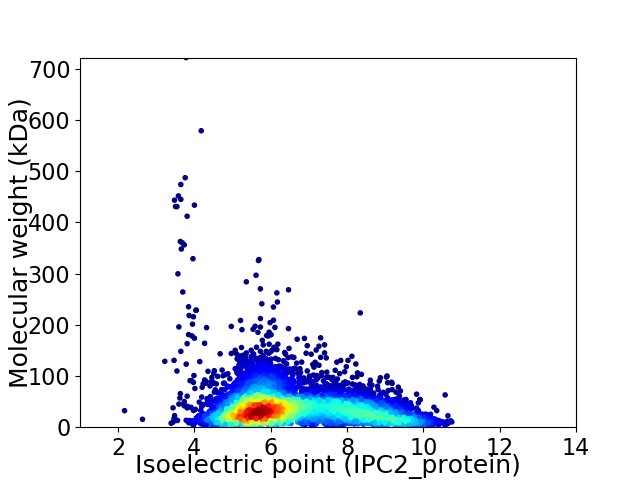
Virtual 2D-PAGE plot for 6977 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
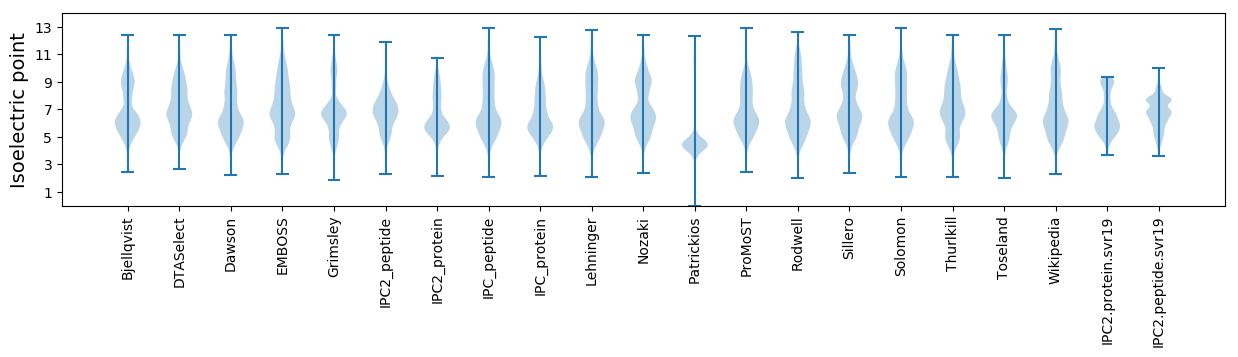
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A517YLX2|A0A517YLX2_9BACT Alpha-D-kanosaminyltransferase OS=Anatilimnocola aggregata OX=2528021 GN=kanE_2 PE=4 SV=1
MM1 pKa = 7.61SRR3 pKa = 11.84INNRR7 pKa = 11.84NRR9 pKa = 11.84NHH11 pKa = 6.62SKK13 pKa = 9.81PAPVPRR19 pKa = 11.84WRR21 pKa = 11.84RR22 pKa = 11.84GVIEE26 pKa = 4.12KK27 pKa = 10.96LEE29 pKa = 4.0ARR31 pKa = 11.84EE32 pKa = 4.15VFSGTPLPVLLVIADD47 pKa = 3.86KK48 pKa = 11.2QDD50 pKa = 3.14FFYY53 pKa = 11.06QEE55 pKa = 3.95YY56 pKa = 10.6GDD58 pKa = 4.49TYY60 pKa = 11.43DD61 pKa = 3.57SLIARR66 pKa = 11.84GVEE69 pKa = 3.96VVVAAEE75 pKa = 4.16TTDD78 pKa = 3.43PSVAHH83 pKa = 7.02PFTGQGAGDD92 pKa = 4.57GIVTPDD98 pKa = 3.01IALADD103 pKa = 3.88VQAEE107 pKa = 4.3NYY109 pKa = 9.82SAIAFVGGWGASMYY123 pKa = 10.5QYY125 pKa = 11.24AFPGDD130 pKa = 4.17YY131 pKa = 11.26ANDD134 pKa = 3.8AYY136 pKa = 10.82DD137 pKa = 4.01GNEE140 pKa = 3.7ATQTIVNNLIGDD152 pKa = 4.99FIDD155 pKa = 3.22QDD157 pKa = 3.7KK158 pKa = 10.76YY159 pKa = 10.87VAAVCNGVTVLAWARR174 pKa = 11.84VDD176 pKa = 3.83GVSPLAGRR184 pKa = 11.84TAAVPYY190 pKa = 9.61IGSPAATYY198 pKa = 9.6MGIEE202 pKa = 4.08YY203 pKa = 10.66GNYY206 pKa = 8.98GWAQPMQMDD215 pKa = 4.36ANGINYY221 pKa = 8.38FPSGAIGVADD231 pKa = 5.04DD232 pKa = 4.71PSDD235 pKa = 4.07DD236 pKa = 3.51VWVDD240 pKa = 3.35GQFITAEE247 pKa = 4.27NNHH250 pKa = 5.89SAAEE254 pKa = 4.06FGDD257 pKa = 4.17VIATHH262 pKa = 6.72VINAAVEE269 pKa = 4.24EE270 pKa = 4.42VEE272 pKa = 4.85EE273 pKa = 4.38VPEE276 pKa = 4.07NQAPIVSPGNLVIAEE291 pKa = 4.07NSVAGTVVGQVIASDD306 pKa = 3.72SDD308 pKa = 3.52LGQLLTYY315 pKa = 10.42AIIGGNDD322 pKa = 2.82SGAFTIEE329 pKa = 3.85AATGVITVANPAALDD344 pKa = 4.01FEE346 pKa = 4.82ASPLFEE352 pKa = 4.47LTVQVTDD359 pKa = 3.63NGTPSLNGVASVMIDD374 pKa = 3.51LLDD377 pKa = 4.04EE378 pKa = 5.17LEE380 pKa = 4.48TPPGPVSHH388 pKa = 7.72DD389 pKa = 3.56GPNVLVQGTADD400 pKa = 3.98ADD402 pKa = 4.11TIYY405 pKa = 10.19IWSDD409 pKa = 3.49YY410 pKa = 10.77YY411 pKa = 11.13GQQFVWINGLQTGPLALGSGGRR433 pKa = 11.84VIVFGGDD440 pKa = 3.3GNDD443 pKa = 3.07QVYY446 pKa = 7.97ATDD449 pKa = 3.37SAKK452 pKa = 10.47PVTIYY457 pKa = 11.32GEE459 pKa = 4.54GGHH462 pKa = 7.27DD463 pKa = 3.72SLTGGSANDD472 pKa = 3.83VIDD475 pKa = 5.14GGEE478 pKa = 3.87GWDD481 pKa = 4.49RR482 pKa = 11.84IWAGGGDD489 pKa = 3.96DD490 pKa = 5.76LIVGGAGTDD499 pKa = 3.24WLHH502 pKa = 5.99GRR504 pKa = 11.84EE505 pKa = 4.32GNDD508 pKa = 4.22LIVGGDD514 pKa = 3.65GDD516 pKa = 4.27DD517 pKa = 3.66YY518 pKa = 11.68LYY520 pKa = 11.26GFTGDD525 pKa = 4.11DD526 pKa = 3.66VLIGGQGSDD535 pKa = 3.48RR536 pKa = 11.84IEE538 pKa = 4.46GEE540 pKa = 3.94SGEE543 pKa = 4.49DD544 pKa = 3.44LLIGGTTSYY553 pKa = 11.43DD554 pKa = 3.68SNQLALEE561 pKa = 5.03AILADD566 pKa = 3.53WTAGADD572 pKa = 3.46LAGRR576 pKa = 11.84SGVFQLTFNGQTTADD591 pKa = 4.4DD592 pKa = 3.8SAQDD596 pKa = 3.8VLIGGADD603 pKa = 3.78NDD605 pKa = 3.94WLLIHH610 pKa = 7.07ASDD613 pKa = 4.37YY614 pKa = 10.7VHH616 pKa = 7.32ASQAGDD622 pKa = 3.66LLDD625 pKa = 3.63VLL627 pKa = 4.93
MM1 pKa = 7.61SRR3 pKa = 11.84INNRR7 pKa = 11.84NRR9 pKa = 11.84NHH11 pKa = 6.62SKK13 pKa = 9.81PAPVPRR19 pKa = 11.84WRR21 pKa = 11.84RR22 pKa = 11.84GVIEE26 pKa = 4.12KK27 pKa = 10.96LEE29 pKa = 4.0ARR31 pKa = 11.84EE32 pKa = 4.15VFSGTPLPVLLVIADD47 pKa = 3.86KK48 pKa = 11.2QDD50 pKa = 3.14FFYY53 pKa = 11.06QEE55 pKa = 3.95YY56 pKa = 10.6GDD58 pKa = 4.49TYY60 pKa = 11.43DD61 pKa = 3.57SLIARR66 pKa = 11.84GVEE69 pKa = 3.96VVVAAEE75 pKa = 4.16TTDD78 pKa = 3.43PSVAHH83 pKa = 7.02PFTGQGAGDD92 pKa = 4.57GIVTPDD98 pKa = 3.01IALADD103 pKa = 3.88VQAEE107 pKa = 4.3NYY109 pKa = 9.82SAIAFVGGWGASMYY123 pKa = 10.5QYY125 pKa = 11.24AFPGDD130 pKa = 4.17YY131 pKa = 11.26ANDD134 pKa = 3.8AYY136 pKa = 10.82DD137 pKa = 4.01GNEE140 pKa = 3.7ATQTIVNNLIGDD152 pKa = 4.99FIDD155 pKa = 3.22QDD157 pKa = 3.7KK158 pKa = 10.76YY159 pKa = 10.87VAAVCNGVTVLAWARR174 pKa = 11.84VDD176 pKa = 3.83GVSPLAGRR184 pKa = 11.84TAAVPYY190 pKa = 9.61IGSPAATYY198 pKa = 9.6MGIEE202 pKa = 4.08YY203 pKa = 10.66GNYY206 pKa = 8.98GWAQPMQMDD215 pKa = 4.36ANGINYY221 pKa = 8.38FPSGAIGVADD231 pKa = 5.04DD232 pKa = 4.71PSDD235 pKa = 4.07DD236 pKa = 3.51VWVDD240 pKa = 3.35GQFITAEE247 pKa = 4.27NNHH250 pKa = 5.89SAAEE254 pKa = 4.06FGDD257 pKa = 4.17VIATHH262 pKa = 6.72VINAAVEE269 pKa = 4.24EE270 pKa = 4.42VEE272 pKa = 4.85EE273 pKa = 4.38VPEE276 pKa = 4.07NQAPIVSPGNLVIAEE291 pKa = 4.07NSVAGTVVGQVIASDD306 pKa = 3.72SDD308 pKa = 3.52LGQLLTYY315 pKa = 10.42AIIGGNDD322 pKa = 2.82SGAFTIEE329 pKa = 3.85AATGVITVANPAALDD344 pKa = 4.01FEE346 pKa = 4.82ASPLFEE352 pKa = 4.47LTVQVTDD359 pKa = 3.63NGTPSLNGVASVMIDD374 pKa = 3.51LLDD377 pKa = 4.04EE378 pKa = 5.17LEE380 pKa = 4.48TPPGPVSHH388 pKa = 7.72DD389 pKa = 3.56GPNVLVQGTADD400 pKa = 3.98ADD402 pKa = 4.11TIYY405 pKa = 10.19IWSDD409 pKa = 3.49YY410 pKa = 10.77YY411 pKa = 11.13GQQFVWINGLQTGPLALGSGGRR433 pKa = 11.84VIVFGGDD440 pKa = 3.3GNDD443 pKa = 3.07QVYY446 pKa = 7.97ATDD449 pKa = 3.37SAKK452 pKa = 10.47PVTIYY457 pKa = 11.32GEE459 pKa = 4.54GGHH462 pKa = 7.27DD463 pKa = 3.72SLTGGSANDD472 pKa = 3.83VIDD475 pKa = 5.14GGEE478 pKa = 3.87GWDD481 pKa = 4.49RR482 pKa = 11.84IWAGGGDD489 pKa = 3.96DD490 pKa = 5.76LIVGGAGTDD499 pKa = 3.24WLHH502 pKa = 5.99GRR504 pKa = 11.84EE505 pKa = 4.32GNDD508 pKa = 4.22LIVGGDD514 pKa = 3.65GDD516 pKa = 4.27DD517 pKa = 3.66YY518 pKa = 11.68LYY520 pKa = 11.26GFTGDD525 pKa = 4.11DD526 pKa = 3.66VLIGGQGSDD535 pKa = 3.48RR536 pKa = 11.84IEE538 pKa = 4.46GEE540 pKa = 3.94SGEE543 pKa = 4.49DD544 pKa = 3.44LLIGGTTSYY553 pKa = 11.43DD554 pKa = 3.68SNQLALEE561 pKa = 5.03AILADD566 pKa = 3.53WTAGADD572 pKa = 3.46LAGRR576 pKa = 11.84SGVFQLTFNGQTTADD591 pKa = 4.4DD592 pKa = 3.8SAQDD596 pKa = 3.8VLIGGADD603 pKa = 3.78NDD605 pKa = 3.94WLLIHH610 pKa = 7.07ASDD613 pKa = 4.37YY614 pKa = 10.7VHH616 pKa = 7.32ASQAGDD622 pKa = 3.66LLDD625 pKa = 3.63VLL627 pKa = 4.93
Molecular weight: 65.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A517YDL6|A0A517YDL6_9BACT SHD1 domain-containing protein OS=Anatilimnocola aggregata OX=2528021 GN=ETAA8_34200 PE=4 SV=1
MM1 pKa = 7.64RR2 pKa = 11.84FDD4 pKa = 3.4AARR7 pKa = 11.84GRR9 pKa = 11.84FGRR12 pKa = 11.84TTKK15 pKa = 10.75DD16 pKa = 3.21RR17 pKa = 11.84LEE19 pKa = 4.38SAADD23 pKa = 3.23QWLSPWCWPPCQMIAAISCWVEE45 pKa = 3.52HH46 pKa = 6.35CFCIVLKK53 pKa = 10.75RR54 pKa = 11.84RR55 pKa = 11.84APLFADD61 pKa = 3.84EE62 pKa = 5.24CARR65 pKa = 11.84QLLAEE70 pKa = 4.25QASKK74 pKa = 10.8RR75 pKa = 11.84ADD77 pKa = 3.54CLSGQRR83 pKa = 11.84WFVRR87 pKa = 11.84VEE89 pKa = 3.77YY90 pKa = 10.67RR91 pKa = 11.84PDD93 pKa = 3.47LQTSAARR100 pKa = 11.84CWRR103 pKa = 11.84VRR105 pKa = 11.84KK106 pKa = 9.53HH107 pKa = 6.44VDD109 pKa = 3.86GISNQFHH116 pKa = 6.7SIQRR120 pKa = 11.84TVFPWKK126 pKa = 10.43FIIDD130 pKa = 4.16LQYY133 pKa = 10.51FAYY136 pKa = 10.72LSGFSLRR143 pKa = 11.84RR144 pKa = 11.84LPARR148 pKa = 11.84IRR150 pKa = 11.84PNSLFIARR158 pKa = 11.84LTRR161 pKa = 11.84RR162 pKa = 11.84SIRR165 pKa = 11.84QPQSCLCEE173 pKa = 4.44STSEE177 pKa = 4.15TSPARR182 pKa = 11.84PWRR185 pKa = 11.84FRR187 pKa = 11.84KK188 pKa = 10.19NSVGSTAPSWFFSQGKK204 pKa = 9.13
MM1 pKa = 7.64RR2 pKa = 11.84FDD4 pKa = 3.4AARR7 pKa = 11.84GRR9 pKa = 11.84FGRR12 pKa = 11.84TTKK15 pKa = 10.75DD16 pKa = 3.21RR17 pKa = 11.84LEE19 pKa = 4.38SAADD23 pKa = 3.23QWLSPWCWPPCQMIAAISCWVEE45 pKa = 3.52HH46 pKa = 6.35CFCIVLKK53 pKa = 10.75RR54 pKa = 11.84RR55 pKa = 11.84APLFADD61 pKa = 3.84EE62 pKa = 5.24CARR65 pKa = 11.84QLLAEE70 pKa = 4.25QASKK74 pKa = 10.8RR75 pKa = 11.84ADD77 pKa = 3.54CLSGQRR83 pKa = 11.84WFVRR87 pKa = 11.84VEE89 pKa = 3.77YY90 pKa = 10.67RR91 pKa = 11.84PDD93 pKa = 3.47LQTSAARR100 pKa = 11.84CWRR103 pKa = 11.84VRR105 pKa = 11.84KK106 pKa = 9.53HH107 pKa = 6.44VDD109 pKa = 3.86GISNQFHH116 pKa = 6.7SIQRR120 pKa = 11.84TVFPWKK126 pKa = 10.43FIIDD130 pKa = 4.16LQYY133 pKa = 10.51FAYY136 pKa = 10.72LSGFSLRR143 pKa = 11.84RR144 pKa = 11.84LPARR148 pKa = 11.84IRR150 pKa = 11.84PNSLFIARR158 pKa = 11.84LTRR161 pKa = 11.84RR162 pKa = 11.84SIRR165 pKa = 11.84QPQSCLCEE173 pKa = 4.44STSEE177 pKa = 4.15TSPARR182 pKa = 11.84PWRR185 pKa = 11.84FRR187 pKa = 11.84KK188 pKa = 10.19NSVGSTAPSWFFSQGKK204 pKa = 9.13
Molecular weight: 23.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2600830 |
29 |
7263 |
372.8 |
40.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.522 ± 0.039 | 1.096 ± 0.014 |
5.477 ± 0.024 | 5.839 ± 0.034 |
3.647 ± 0.016 | 7.907 ± 0.052 |
2.165 ± 0.019 | 4.719 ± 0.023 |
4.186 ± 0.036 | 10.285 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.976 ± 0.013 | 3.314 ± 0.035 |
5.562 ± 0.03 | 4.273 ± 0.025 |
6.35 ± 0.041 | 5.929 ± 0.027 |
5.763 ± 0.055 | 7.072 ± 0.024 |
1.545 ± 0.016 | 2.374 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
