
Bovine polyomavirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; unclassified Polyomaviridae
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
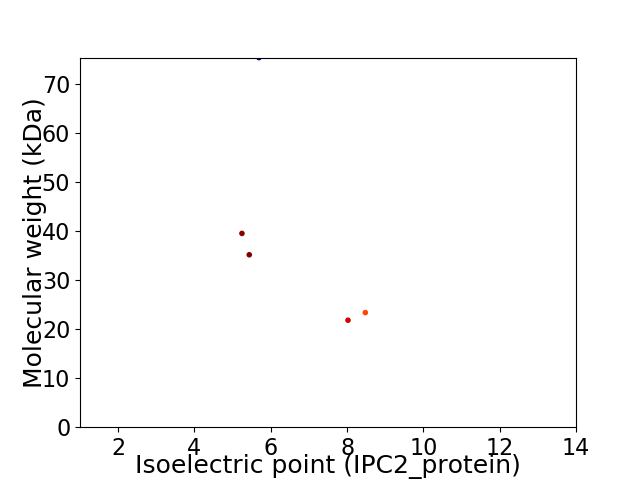
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A097IQX9|A0A097IQX9_9POLY VP3 OS=Bovine polyomavirus 3 OX=1561705 PE=4 SV=1
MM1 pKa = 6.89ATCGAGRR8 pKa = 11.84GKK10 pKa = 9.05VTLPRR15 pKa = 11.84QIKK18 pKa = 10.05KK19 pKa = 10.72GGAEE23 pKa = 4.08VLDD26 pKa = 4.02TVPLSEE32 pKa = 4.14TTQYY36 pKa = 11.58AVEE39 pKa = 4.6FVNKK43 pKa = 8.67PTFLKK48 pKa = 10.4TNGDD52 pKa = 3.21PEE54 pKa = 4.14YY55 pKa = 10.47RR56 pKa = 11.84SRR58 pKa = 11.84GLQYY62 pKa = 11.31LLDD65 pKa = 4.92DD66 pKa = 4.21PTVGVQGALCYY77 pKa = 10.45SLGVIDD83 pKa = 6.32LPEE86 pKa = 5.16IPNAMCEE93 pKa = 4.01DD94 pKa = 3.94SVIVWEE100 pKa = 4.62AYY102 pKa = 9.68RR103 pKa = 11.84VEE105 pKa = 4.39TEE107 pKa = 5.17LITLPVLGSGGYY119 pKa = 9.66VSATKK124 pKa = 10.01TIASVEE130 pKa = 4.18GPQMYY135 pKa = 9.41FWAVGGSPLDD145 pKa = 3.48VVGFNPDD152 pKa = 3.2PDD154 pKa = 3.74GTYY157 pKa = 10.38TFAAEE162 pKa = 3.97LAAPKK167 pKa = 9.42GTISVAGPGATKK179 pKa = 10.59SQVTTPKK186 pKa = 9.44YY187 pKa = 9.13TVEE190 pKa = 3.77GWAADD195 pKa = 3.47PSMNDD200 pKa = 2.7NTKK203 pKa = 10.03YY204 pKa = 9.78FGRR207 pKa = 11.84IIGGSQTPPVITYY220 pKa = 10.38GNGSTTPLVDD230 pKa = 3.75EE231 pKa = 4.82YY232 pKa = 11.66GVGPLCLHH240 pKa = 6.21GVCYY244 pKa = 9.1LTSVDD249 pKa = 4.92LMGTVGWPGQATLPDD264 pKa = 3.71GGVKK268 pKa = 9.75KK269 pKa = 10.46RR270 pKa = 11.84ALQTAAGRR278 pKa = 11.84FFRR281 pKa = 11.84VHH283 pKa = 5.22FRR285 pKa = 11.84QRR287 pKa = 11.84RR288 pKa = 11.84VKK290 pKa = 10.76NPWTMEE296 pKa = 3.7QMLSQFLRR304 pKa = 11.84PKK306 pKa = 10.58APVVTGAQTEE316 pKa = 4.4VSEE319 pKa = 4.51VTMTQDD325 pKa = 3.84KK326 pKa = 10.44GPASIPPTLEE336 pKa = 3.55ASVGYY341 pKa = 9.44GQAIFNVVDD350 pKa = 4.05GQLVYY355 pKa = 10.59PDD357 pKa = 4.03GQGGSINIGTRR368 pKa = 11.84TQMM371 pKa = 3.74
MM1 pKa = 6.89ATCGAGRR8 pKa = 11.84GKK10 pKa = 9.05VTLPRR15 pKa = 11.84QIKK18 pKa = 10.05KK19 pKa = 10.72GGAEE23 pKa = 4.08VLDD26 pKa = 4.02TVPLSEE32 pKa = 4.14TTQYY36 pKa = 11.58AVEE39 pKa = 4.6FVNKK43 pKa = 8.67PTFLKK48 pKa = 10.4TNGDD52 pKa = 3.21PEE54 pKa = 4.14YY55 pKa = 10.47RR56 pKa = 11.84SRR58 pKa = 11.84GLQYY62 pKa = 11.31LLDD65 pKa = 4.92DD66 pKa = 4.21PTVGVQGALCYY77 pKa = 10.45SLGVIDD83 pKa = 6.32LPEE86 pKa = 5.16IPNAMCEE93 pKa = 4.01DD94 pKa = 3.94SVIVWEE100 pKa = 4.62AYY102 pKa = 9.68RR103 pKa = 11.84VEE105 pKa = 4.39TEE107 pKa = 5.17LITLPVLGSGGYY119 pKa = 9.66VSATKK124 pKa = 10.01TIASVEE130 pKa = 4.18GPQMYY135 pKa = 9.41FWAVGGSPLDD145 pKa = 3.48VVGFNPDD152 pKa = 3.2PDD154 pKa = 3.74GTYY157 pKa = 10.38TFAAEE162 pKa = 3.97LAAPKK167 pKa = 9.42GTISVAGPGATKK179 pKa = 10.59SQVTTPKK186 pKa = 9.44YY187 pKa = 9.13TVEE190 pKa = 3.77GWAADD195 pKa = 3.47PSMNDD200 pKa = 2.7NTKK203 pKa = 10.03YY204 pKa = 9.78FGRR207 pKa = 11.84IIGGSQTPPVITYY220 pKa = 10.38GNGSTTPLVDD230 pKa = 3.75EE231 pKa = 4.82YY232 pKa = 11.66GVGPLCLHH240 pKa = 6.21GVCYY244 pKa = 9.1LTSVDD249 pKa = 4.92LMGTVGWPGQATLPDD264 pKa = 3.71GGVKK268 pKa = 9.75KK269 pKa = 10.46RR270 pKa = 11.84ALQTAAGRR278 pKa = 11.84FFRR281 pKa = 11.84VHH283 pKa = 5.22FRR285 pKa = 11.84QRR287 pKa = 11.84RR288 pKa = 11.84VKK290 pKa = 10.76NPWTMEE296 pKa = 3.7QMLSQFLRR304 pKa = 11.84PKK306 pKa = 10.58APVVTGAQTEE316 pKa = 4.4VSEE319 pKa = 4.51VTMTQDD325 pKa = 3.84KK326 pKa = 10.44GPASIPPTLEE336 pKa = 3.55ASVGYY341 pKa = 9.44GQAIFNVVDD350 pKa = 4.05GQLVYY355 pKa = 10.59PDD357 pKa = 4.03GQGGSINIGTRR368 pKa = 11.84TQMM371 pKa = 3.74
Molecular weight: 39.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A097IR00|A0A097IR00_9POLY VP2 OS=Bovine polyomavirus 3 OX=1561705 PE=4 SV=1
MM1 pKa = 7.76ALQLWFPQTGLWQNYY16 pKa = 7.8GVSLPDD22 pKa = 2.84WWSSLLSQLPDD33 pKa = 4.1PYY35 pKa = 10.87TILQDD40 pKa = 3.19IARR43 pKa = 11.84GIWTSYY49 pKa = 9.04YY50 pKa = 10.17RR51 pKa = 11.84AGQEE55 pKa = 3.58IVRR58 pKa = 11.84RR59 pKa = 11.84TVGNQIRR66 pKa = 11.84GFVGSVQEE74 pKa = 4.51NLLQTGRR81 pKa = 11.84AIVEE85 pKa = 4.11RR86 pKa = 11.84APDD89 pKa = 3.97PVTGLINVWNWATEE103 pKa = 3.91HH104 pKa = 4.96QRR106 pKa = 11.84QWEE109 pKa = 4.34TQALLEE115 pKa = 4.25GQPVFGRR122 pKa = 11.84EE123 pKa = 4.28TQWEE127 pKa = 4.46SQNLPIDD134 pKa = 3.84GGNNQRR140 pKa = 11.84DD141 pKa = 3.57GWHH144 pKa = 6.89WGGQWVTMPGSNLGAYY160 pKa = 7.4TVPQWMLYY168 pKa = 9.5VLEE171 pKa = 4.49EE172 pKa = 4.2VEE174 pKa = 4.54KK175 pKa = 11.14DD176 pKa = 3.28LTAHH180 pKa = 6.57HH181 pKa = 6.71GNMRR185 pKa = 11.84SRR187 pKa = 11.84KK188 pKa = 9.01RR189 pKa = 11.84QGDD192 pKa = 3.45PAKK195 pKa = 9.75TNKK198 pKa = 9.16KK199 pKa = 9.56RR200 pKa = 11.84RR201 pKa = 11.84RR202 pKa = 3.59
MM1 pKa = 7.76ALQLWFPQTGLWQNYY16 pKa = 7.8GVSLPDD22 pKa = 2.84WWSSLLSQLPDD33 pKa = 4.1PYY35 pKa = 10.87TILQDD40 pKa = 3.19IARR43 pKa = 11.84GIWTSYY49 pKa = 9.04YY50 pKa = 10.17RR51 pKa = 11.84AGQEE55 pKa = 3.58IVRR58 pKa = 11.84RR59 pKa = 11.84TVGNQIRR66 pKa = 11.84GFVGSVQEE74 pKa = 4.51NLLQTGRR81 pKa = 11.84AIVEE85 pKa = 4.11RR86 pKa = 11.84APDD89 pKa = 3.97PVTGLINVWNWATEE103 pKa = 3.91HH104 pKa = 4.96QRR106 pKa = 11.84QWEE109 pKa = 4.34TQALLEE115 pKa = 4.25GQPVFGRR122 pKa = 11.84EE123 pKa = 4.28TQWEE127 pKa = 4.46SQNLPIDD134 pKa = 3.84GGNNQRR140 pKa = 11.84DD141 pKa = 3.57GWHH144 pKa = 6.89WGGQWVTMPGSNLGAYY160 pKa = 7.4TVPQWMLYY168 pKa = 9.5VLEE171 pKa = 4.49EE172 pKa = 4.2VEE174 pKa = 4.54KK175 pKa = 11.14DD176 pKa = 3.28LTAHH180 pKa = 6.57HH181 pKa = 6.71GNMRR185 pKa = 11.84SRR187 pKa = 11.84KK188 pKa = 9.01RR189 pKa = 11.84QGDD192 pKa = 3.45PAKK195 pKa = 9.75TNKK198 pKa = 9.16KK199 pKa = 9.56RR200 pKa = 11.84RR201 pKa = 11.84RR202 pKa = 3.59
Molecular weight: 23.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1756 |
188 |
673 |
351.2 |
39.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.403 ± 0.753 | 1.993 ± 0.637 |
4.613 ± 0.48 | 6.15 ± 0.576 |
3.417 ± 0.481 | 9.282 ± 1.099 |
1.936 ± 0.419 | 4.328 ± 0.275 |
5.41 ± 1.151 | 8.827 ± 0.564 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.278 ± 0.205 | 4.271 ± 0.474 |
5.979 ± 0.413 | 5.125 ± 0.756 |
4.67 ± 0.698 | 5.011 ± 0.112 |
6.549 ± 0.819 | 6.891 ± 0.736 |
3.303 ± 0.78 | 2.563 ± 0.444 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
