
Xinzhou partiti-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
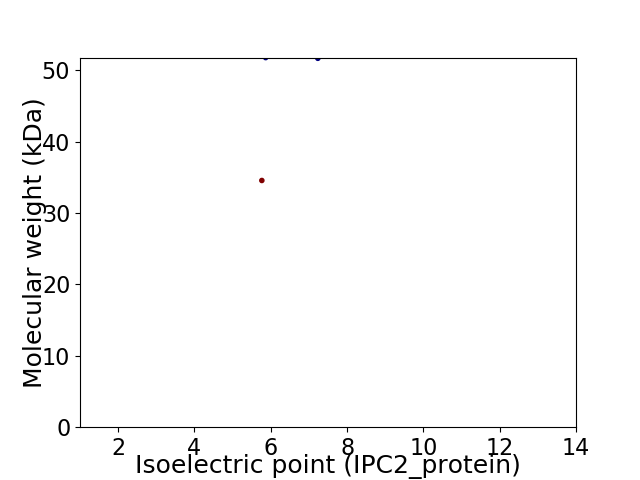
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLW6|A0A1L3KLW6_9VIRU RdRp OS=Xinzhou partiti-like virus 1 OX=1923776 PE=4 SV=1
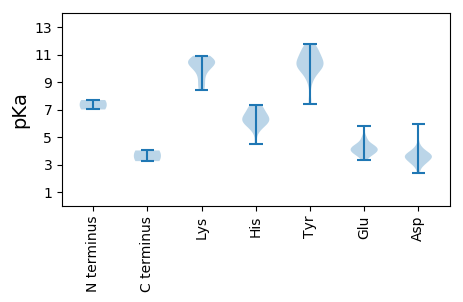
MM1 pKa = 7.69SKK3 pKa = 10.44RR4 pKa = 11.84PAEE7 pKa = 4.09PAFGTPAKK15 pKa = 10.79VMAGDD20 pKa = 3.58SADD23 pKa = 3.08AHH25 pKa = 5.88TEE27 pKa = 3.75VSGVIGQDD35 pKa = 3.12NPVPAPRR42 pKa = 11.84RR43 pKa = 11.84QSANKK48 pKa = 9.04QQLLDD53 pKa = 3.81SEE55 pKa = 4.71LASRR59 pKa = 11.84PKK61 pKa = 10.45PEE63 pKa = 4.16EE64 pKa = 3.71LNILRR69 pKa = 11.84TYY71 pKa = 10.23ASNVIGPTPCHH82 pKa = 6.26SDD84 pKa = 2.55IYY86 pKa = 11.25VLYY89 pKa = 9.49THH91 pKa = 7.16TALDD95 pKa = 3.71AFAKK99 pKa = 10.11LVRR102 pKa = 11.84RR103 pKa = 11.84QILNTLYY110 pKa = 10.13PDD112 pKa = 4.6GDD114 pKa = 4.07FLPQGVCSEE123 pKa = 4.37EE124 pKa = 3.89DD125 pKa = 3.1WVIVVQYY132 pKa = 10.01ILKK135 pKa = 10.41SRR137 pKa = 11.84IDD139 pKa = 3.68LVYY142 pKa = 10.81ANYY145 pKa = 10.24SGRR148 pKa = 11.84RR149 pKa = 11.84PPDD152 pKa = 3.1RR153 pKa = 11.84VPLVRR158 pKa = 11.84IVMPRR163 pKa = 11.84SIASLINGIGVKK175 pKa = 9.31QVFHH179 pKa = 7.11GNFSVCPQPGDD190 pKa = 3.68PPVDD194 pKa = 3.26RR195 pKa = 11.84ATWLINLATHH205 pKa = 7.05ARR207 pKa = 11.84IQSFSNLVLAALQRR221 pKa = 11.84GVIHH225 pKa = 7.31PGTASNLVEE234 pKa = 4.17GTGWWLLYY242 pKa = 10.25AANSTNRR249 pKa = 11.84GQIATIDD256 pKa = 3.97TQSVTVWAQFPEE268 pKa = 4.36WTPSDD273 pKa = 3.76GLLCAIAATGFIGTFIDD290 pKa = 4.14DD291 pKa = 3.58WTEE294 pKa = 3.58YY295 pKa = 9.47TYY297 pKa = 11.49KK298 pKa = 10.7LEE300 pKa = 4.19SVTDD304 pKa = 3.95SVGMRR309 pKa = 11.84ASYY312 pKa = 10.89AVLGG316 pKa = 4.02
MM1 pKa = 7.69SKK3 pKa = 10.44RR4 pKa = 11.84PAEE7 pKa = 4.09PAFGTPAKK15 pKa = 10.79VMAGDD20 pKa = 3.58SADD23 pKa = 3.08AHH25 pKa = 5.88TEE27 pKa = 3.75VSGVIGQDD35 pKa = 3.12NPVPAPRR42 pKa = 11.84RR43 pKa = 11.84QSANKK48 pKa = 9.04QQLLDD53 pKa = 3.81SEE55 pKa = 4.71LASRR59 pKa = 11.84PKK61 pKa = 10.45PEE63 pKa = 4.16EE64 pKa = 3.71LNILRR69 pKa = 11.84TYY71 pKa = 10.23ASNVIGPTPCHH82 pKa = 6.26SDD84 pKa = 2.55IYY86 pKa = 11.25VLYY89 pKa = 9.49THH91 pKa = 7.16TALDD95 pKa = 3.71AFAKK99 pKa = 10.11LVRR102 pKa = 11.84RR103 pKa = 11.84QILNTLYY110 pKa = 10.13PDD112 pKa = 4.6GDD114 pKa = 4.07FLPQGVCSEE123 pKa = 4.37EE124 pKa = 3.89DD125 pKa = 3.1WVIVVQYY132 pKa = 10.01ILKK135 pKa = 10.41SRR137 pKa = 11.84IDD139 pKa = 3.68LVYY142 pKa = 10.81ANYY145 pKa = 10.24SGRR148 pKa = 11.84RR149 pKa = 11.84PPDD152 pKa = 3.1RR153 pKa = 11.84VPLVRR158 pKa = 11.84IVMPRR163 pKa = 11.84SIASLINGIGVKK175 pKa = 9.31QVFHH179 pKa = 7.11GNFSVCPQPGDD190 pKa = 3.68PPVDD194 pKa = 3.26RR195 pKa = 11.84ATWLINLATHH205 pKa = 7.05ARR207 pKa = 11.84IQSFSNLVLAALQRR221 pKa = 11.84GVIHH225 pKa = 7.31PGTASNLVEE234 pKa = 4.17GTGWWLLYY242 pKa = 10.25AANSTNRR249 pKa = 11.84GQIATIDD256 pKa = 3.97TQSVTVWAQFPEE268 pKa = 4.36WTPSDD273 pKa = 3.76GLLCAIAATGFIGTFIDD290 pKa = 4.14DD291 pKa = 3.58WTEE294 pKa = 3.58YY295 pKa = 9.47TYY297 pKa = 11.49KK298 pKa = 10.7LEE300 pKa = 4.19SVTDD304 pKa = 3.95SVGMRR309 pKa = 11.84ASYY312 pKa = 10.89AVLGG316 pKa = 4.02
Molecular weight: 34.55 kDa
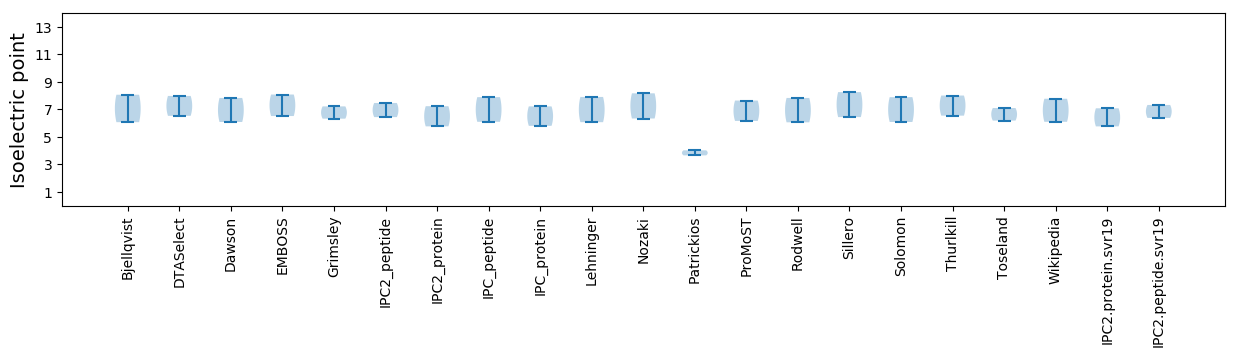
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLW6|A0A1L3KLW6_9VIRU RdRp OS=Xinzhou partiti-like virus 1 OX=1923776 PE=4 SV=1
MM1 pKa = 7.0STLEE5 pKa = 4.42FVRR8 pKa = 11.84QLPSYY13 pKa = 11.39SLMQHH18 pKa = 5.97GTPCDD23 pKa = 3.51LYY25 pKa = 11.57ARR27 pKa = 11.84GLINALYY34 pKa = 9.96PEE36 pKa = 4.99ALPSIDD42 pKa = 3.47EE43 pKa = 4.35YY44 pKa = 11.62SRR46 pKa = 11.84GMYY49 pKa = 10.07DD50 pKa = 3.58IEE52 pKa = 3.98HH53 pKa = 7.33HH54 pKa = 5.42YY55 pKa = 8.88TAFYY59 pKa = 10.09KK60 pKa = 10.56YY61 pKa = 10.54SRR63 pKa = 11.84PQYY66 pKa = 9.9LEE68 pKa = 3.33QAIKK72 pKa = 10.45HH73 pKa = 5.56RR74 pKa = 11.84LEE76 pKa = 4.1SSSLIKK82 pKa = 9.82EE83 pKa = 3.51ARR85 pKa = 11.84AIVEE89 pKa = 4.3SEE91 pKa = 3.9LTEE94 pKa = 4.31SFTVQAISMSHH105 pKa = 6.58LDD107 pKa = 3.19AVSFVGSSAAGYY119 pKa = 10.18GYY121 pKa = 10.9VGNKK125 pKa = 8.47RR126 pKa = 11.84DD127 pKa = 3.48NYY129 pKa = 10.51LVARR133 pKa = 11.84GHH135 pKa = 5.91ATSNLANFRR144 pKa = 11.84RR145 pKa = 11.84WGSKK149 pKa = 9.94LRR151 pKa = 11.84LTPYY155 pKa = 9.65KK156 pKa = 10.42AYY158 pKa = 10.92SRR160 pKa = 11.84TQLALRR166 pKa = 11.84ADD168 pKa = 3.67PKK170 pKa = 10.53IRR172 pKa = 11.84HH173 pKa = 4.48VWGAPFHH180 pKa = 6.5TILIEE185 pKa = 4.02GTIAQPIIVNLQLYY199 pKa = 7.38DD200 pKa = 3.5QPIYY204 pKa = 10.2IGKK207 pKa = 10.09DD208 pKa = 3.05MFKK211 pKa = 10.45EE212 pKa = 3.93LPYY215 pKa = 10.33TLNRR219 pKa = 11.84LLRR222 pKa = 11.84DD223 pKa = 3.4NFFAYY228 pKa = 10.55CIDD231 pKa = 3.14ISAFDD236 pKa = 3.58SSVNQIFIDD245 pKa = 3.52WFFDD249 pKa = 4.2FVSKK253 pKa = 9.64TVTFPNAFTRR263 pKa = 11.84SAVDD267 pKa = 2.94YY268 pKa = 11.15CRR270 pKa = 11.84FEE272 pKa = 5.0LKK274 pKa = 8.8HH275 pKa = 6.07TPVVMPDD282 pKa = 2.38GKK284 pKa = 10.46MYY286 pKa = 10.16ICHH289 pKa = 6.61TGIPSGSYY297 pKa = 7.66FTQLIDD303 pKa = 3.74SYY305 pKa = 11.79VNLIMLRR312 pKa = 11.84ACQLSILEE320 pKa = 4.61SVVPTYY326 pKa = 11.45VLGDD330 pKa = 3.51DD331 pKa = 5.96SIFCHH336 pKa = 6.53HH337 pKa = 6.73NPNHH341 pKa = 6.28LEE343 pKa = 3.91SFRR346 pKa = 11.84VLFEE350 pKa = 3.9EE351 pKa = 5.81FGFKK355 pKa = 10.91LNVRR359 pKa = 11.84KK360 pKa = 8.38TIVSKK365 pKa = 10.82SSQEE369 pKa = 3.83IVFLGHH375 pKa = 6.17NFYY378 pKa = 11.08GSRR381 pKa = 11.84LTRR384 pKa = 11.84DD385 pKa = 4.05DD386 pKa = 3.59FTLACLAIHH395 pKa = 6.58TEE397 pKa = 4.25TPIVNSSQTVVRR409 pKa = 11.84LASLLYY415 pKa = 10.61DD416 pKa = 3.64SGHH419 pKa = 6.14NSFFLMNLLTAAIKK433 pKa = 10.27RR434 pKa = 11.84YY435 pKa = 9.18GSPRR439 pKa = 11.84PLHH442 pKa = 6.08PPYY445 pKa = 10.65VQLFMLSS452 pKa = 3.25
MM1 pKa = 7.0STLEE5 pKa = 4.42FVRR8 pKa = 11.84QLPSYY13 pKa = 11.39SLMQHH18 pKa = 5.97GTPCDD23 pKa = 3.51LYY25 pKa = 11.57ARR27 pKa = 11.84GLINALYY34 pKa = 9.96PEE36 pKa = 4.99ALPSIDD42 pKa = 3.47EE43 pKa = 4.35YY44 pKa = 11.62SRR46 pKa = 11.84GMYY49 pKa = 10.07DD50 pKa = 3.58IEE52 pKa = 3.98HH53 pKa = 7.33HH54 pKa = 5.42YY55 pKa = 8.88TAFYY59 pKa = 10.09KK60 pKa = 10.56YY61 pKa = 10.54SRR63 pKa = 11.84PQYY66 pKa = 9.9LEE68 pKa = 3.33QAIKK72 pKa = 10.45HH73 pKa = 5.56RR74 pKa = 11.84LEE76 pKa = 4.1SSSLIKK82 pKa = 9.82EE83 pKa = 3.51ARR85 pKa = 11.84AIVEE89 pKa = 4.3SEE91 pKa = 3.9LTEE94 pKa = 4.31SFTVQAISMSHH105 pKa = 6.58LDD107 pKa = 3.19AVSFVGSSAAGYY119 pKa = 10.18GYY121 pKa = 10.9VGNKK125 pKa = 8.47RR126 pKa = 11.84DD127 pKa = 3.48NYY129 pKa = 10.51LVARR133 pKa = 11.84GHH135 pKa = 5.91ATSNLANFRR144 pKa = 11.84RR145 pKa = 11.84WGSKK149 pKa = 9.94LRR151 pKa = 11.84LTPYY155 pKa = 9.65KK156 pKa = 10.42AYY158 pKa = 10.92SRR160 pKa = 11.84TQLALRR166 pKa = 11.84ADD168 pKa = 3.67PKK170 pKa = 10.53IRR172 pKa = 11.84HH173 pKa = 4.48VWGAPFHH180 pKa = 6.5TILIEE185 pKa = 4.02GTIAQPIIVNLQLYY199 pKa = 7.38DD200 pKa = 3.5QPIYY204 pKa = 10.2IGKK207 pKa = 10.09DD208 pKa = 3.05MFKK211 pKa = 10.45EE212 pKa = 3.93LPYY215 pKa = 10.33TLNRR219 pKa = 11.84LLRR222 pKa = 11.84DD223 pKa = 3.4NFFAYY228 pKa = 10.55CIDD231 pKa = 3.14ISAFDD236 pKa = 3.58SSVNQIFIDD245 pKa = 3.52WFFDD249 pKa = 4.2FVSKK253 pKa = 9.64TVTFPNAFTRR263 pKa = 11.84SAVDD267 pKa = 2.94YY268 pKa = 11.15CRR270 pKa = 11.84FEE272 pKa = 5.0LKK274 pKa = 8.8HH275 pKa = 6.07TPVVMPDD282 pKa = 2.38GKK284 pKa = 10.46MYY286 pKa = 10.16ICHH289 pKa = 6.61TGIPSGSYY297 pKa = 7.66FTQLIDD303 pKa = 3.74SYY305 pKa = 11.79VNLIMLRR312 pKa = 11.84ACQLSILEE320 pKa = 4.61SVVPTYY326 pKa = 11.45VLGDD330 pKa = 3.51DD331 pKa = 5.96SIFCHH336 pKa = 6.53HH337 pKa = 6.73NPNHH341 pKa = 6.28LEE343 pKa = 3.91SFRR346 pKa = 11.84VLFEE350 pKa = 3.9EE351 pKa = 5.81FGFKK355 pKa = 10.91LNVRR359 pKa = 11.84KK360 pKa = 8.38TIVSKK365 pKa = 10.82SSQEE369 pKa = 3.83IVFLGHH375 pKa = 6.17NFYY378 pKa = 11.08GSRR381 pKa = 11.84LTRR384 pKa = 11.84DD385 pKa = 4.05DD386 pKa = 3.59FTLACLAIHH395 pKa = 6.58TEE397 pKa = 4.25TPIVNSSQTVVRR409 pKa = 11.84LASLLYY415 pKa = 10.61DD416 pKa = 3.64SGHH419 pKa = 6.14NSFFLMNLLTAAIKK433 pKa = 10.27RR434 pKa = 11.84YY435 pKa = 9.18GSPRR439 pKa = 11.84PLHH442 pKa = 6.08PPYY445 pKa = 10.65VQLFMLSS452 pKa = 3.25
Molecular weight: 51.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
768 |
316 |
452 |
384.0 |
43.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.682 ± 1.133 | 1.432 ± 0.104 |
5.078 ± 0.387 | 3.906 ± 0.266 |
4.948 ± 1.313 | 5.599 ± 0.852 |
2.995 ± 0.685 | 6.51 ± 0.113 |
3.125 ± 0.371 | 9.766 ± 0.764 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.823 ± 0.348 | 4.036 ± 0.048 |
5.859 ± 0.888 | 3.776 ± 0.409 |
5.729 ± 0.021 | 8.464 ± 0.741 |
5.99 ± 0.41 | 7.031 ± 0.946 |
1.302 ± 0.571 | 4.948 ± 0.917 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
