
Hainan black-spectacled toad dimarhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
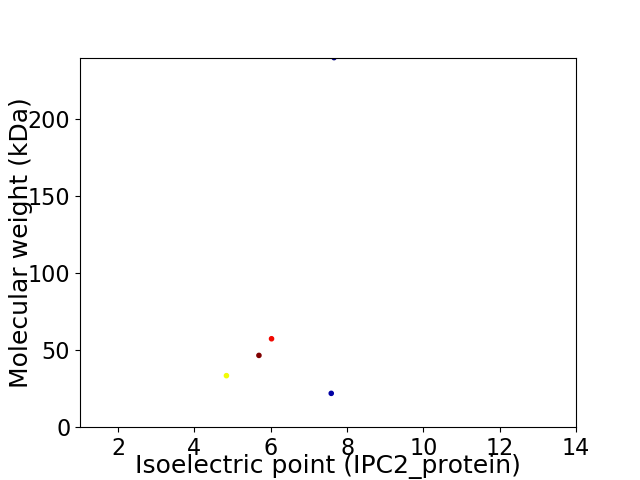
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1GNA1|A0A2P1GNA1_9RHAB Nucleocapsid protein OS=Hainan black-spectacled toad dimarhabdovirus OX=2116358 PE=4 SV=1
MM1 pKa = 7.41SCLNKK6 pKa = 10.29RR7 pKa = 11.84FMKK10 pKa = 9.03KK11 pKa = 7.38TNRR14 pKa = 11.84DD15 pKa = 3.12NKK17 pKa = 10.01MEE19 pKa = 4.42LSNFQNLFPKK29 pKa = 10.26KK30 pKa = 9.88LLSAAVAMEE39 pKa = 4.28EE40 pKa = 4.08KK41 pKa = 9.92HH42 pKa = 6.68HH43 pKa = 6.6KK44 pKa = 10.33SSLPYY49 pKa = 10.11EE50 pKa = 4.44VINPEE55 pKa = 3.61EE56 pKa = 4.14RR57 pKa = 11.84EE58 pKa = 3.89PAKK61 pKa = 11.08VEE63 pKa = 4.08VEE65 pKa = 4.19VPEE68 pKa = 4.28TNKK71 pKa = 10.66DD72 pKa = 3.8FVPQPPEE79 pKa = 3.61KK80 pKa = 10.75ARR82 pKa = 11.84VEE84 pKa = 4.13QYY86 pKa = 11.26LGISLPKK93 pKa = 8.37TQRR96 pKa = 11.84DD97 pKa = 3.24LDD99 pKa = 4.09EE100 pKa = 4.48KK101 pKa = 11.09VKK103 pKa = 10.71EE104 pKa = 4.03LVLSILEE111 pKa = 4.47TYY113 pKa = 9.6GVNHH117 pKa = 7.08AWIKK121 pKa = 10.3VDD123 pKa = 3.8KK124 pKa = 10.82DD125 pKa = 3.15WITFQEE131 pKa = 3.75IDD133 pKa = 3.5EE134 pKa = 5.06DD135 pKa = 4.84GEE137 pKa = 4.07ALEE140 pKa = 5.1ISSSDD145 pKa = 3.52DD146 pKa = 3.43EE147 pKa = 4.68YY148 pKa = 11.6EE149 pKa = 3.84EE150 pKa = 6.5DD151 pKa = 5.55EE152 pKa = 5.23DD153 pKa = 6.26DD154 pKa = 3.83EE155 pKa = 4.82TNEE158 pKa = 4.04SDD160 pKa = 5.12EE161 pKa = 4.76SGSDD165 pKa = 3.32QKK167 pKa = 11.75GEE169 pKa = 4.05AFLQVLRR176 pKa = 11.84PEE178 pKa = 4.09QDD180 pKa = 2.77QGKK183 pKa = 10.1LDD185 pKa = 4.12FKK187 pKa = 10.98NSPSNPFASEE197 pKa = 3.87LNKK200 pKa = 9.92IEE202 pKa = 4.69KK203 pKa = 10.1DD204 pKa = 3.39LPTVLTRR211 pKa = 11.84SQVFFTQLKK220 pKa = 9.98NGFEE224 pKa = 4.36LVSISGSRR232 pKa = 11.84LWVDD236 pKa = 3.84FEE238 pKa = 4.07KK239 pKa = 11.12LKK241 pKa = 10.79IGMHH245 pKa = 6.96EE246 pKa = 3.83ILDD249 pKa = 3.75FPFRR253 pKa = 11.84EE254 pKa = 4.41DD255 pKa = 3.0WSKK258 pKa = 9.88TKK260 pKa = 10.7CLFEE264 pKa = 3.99YY265 pKa = 10.41CKK267 pKa = 10.65KK268 pKa = 10.75NKK270 pKa = 8.35TSRR273 pKa = 11.84AKK275 pKa = 10.7LGMANWGEE283 pKa = 4.33SLRR286 pKa = 11.84VDD288 pKa = 3.69PP289 pKa = 6.04
MM1 pKa = 7.41SCLNKK6 pKa = 10.29RR7 pKa = 11.84FMKK10 pKa = 9.03KK11 pKa = 7.38TNRR14 pKa = 11.84DD15 pKa = 3.12NKK17 pKa = 10.01MEE19 pKa = 4.42LSNFQNLFPKK29 pKa = 10.26KK30 pKa = 9.88LLSAAVAMEE39 pKa = 4.28EE40 pKa = 4.08KK41 pKa = 9.92HH42 pKa = 6.68HH43 pKa = 6.6KK44 pKa = 10.33SSLPYY49 pKa = 10.11EE50 pKa = 4.44VINPEE55 pKa = 3.61EE56 pKa = 4.14RR57 pKa = 11.84EE58 pKa = 3.89PAKK61 pKa = 11.08VEE63 pKa = 4.08VEE65 pKa = 4.19VPEE68 pKa = 4.28TNKK71 pKa = 10.66DD72 pKa = 3.8FVPQPPEE79 pKa = 3.61KK80 pKa = 10.75ARR82 pKa = 11.84VEE84 pKa = 4.13QYY86 pKa = 11.26LGISLPKK93 pKa = 8.37TQRR96 pKa = 11.84DD97 pKa = 3.24LDD99 pKa = 4.09EE100 pKa = 4.48KK101 pKa = 11.09VKK103 pKa = 10.71EE104 pKa = 4.03LVLSILEE111 pKa = 4.47TYY113 pKa = 9.6GVNHH117 pKa = 7.08AWIKK121 pKa = 10.3VDD123 pKa = 3.8KK124 pKa = 10.82DD125 pKa = 3.15WITFQEE131 pKa = 3.75IDD133 pKa = 3.5EE134 pKa = 5.06DD135 pKa = 4.84GEE137 pKa = 4.07ALEE140 pKa = 5.1ISSSDD145 pKa = 3.52DD146 pKa = 3.43EE147 pKa = 4.68YY148 pKa = 11.6EE149 pKa = 3.84EE150 pKa = 6.5DD151 pKa = 5.55EE152 pKa = 5.23DD153 pKa = 6.26DD154 pKa = 3.83EE155 pKa = 4.82TNEE158 pKa = 4.04SDD160 pKa = 5.12EE161 pKa = 4.76SGSDD165 pKa = 3.32QKK167 pKa = 11.75GEE169 pKa = 4.05AFLQVLRR176 pKa = 11.84PEE178 pKa = 4.09QDD180 pKa = 2.77QGKK183 pKa = 10.1LDD185 pKa = 4.12FKK187 pKa = 10.98NSPSNPFASEE197 pKa = 3.87LNKK200 pKa = 9.92IEE202 pKa = 4.69KK203 pKa = 10.1DD204 pKa = 3.39LPTVLTRR211 pKa = 11.84SQVFFTQLKK220 pKa = 9.98NGFEE224 pKa = 4.36LVSISGSRR232 pKa = 11.84LWVDD236 pKa = 3.84FEE238 pKa = 4.07KK239 pKa = 11.12LKK241 pKa = 10.79IGMHH245 pKa = 6.96EE246 pKa = 3.83ILDD249 pKa = 3.75FPFRR253 pKa = 11.84EE254 pKa = 4.41DD255 pKa = 3.0WSKK258 pKa = 9.88TKK260 pKa = 10.7CLFEE264 pKa = 3.99YY265 pKa = 10.41CKK267 pKa = 10.65KK268 pKa = 10.75NKK270 pKa = 8.35TSRR273 pKa = 11.84AKK275 pKa = 10.7LGMANWGEE283 pKa = 4.33SLRR286 pKa = 11.84VDD288 pKa = 3.69PP289 pKa = 6.04
Molecular weight: 33.45 kDa
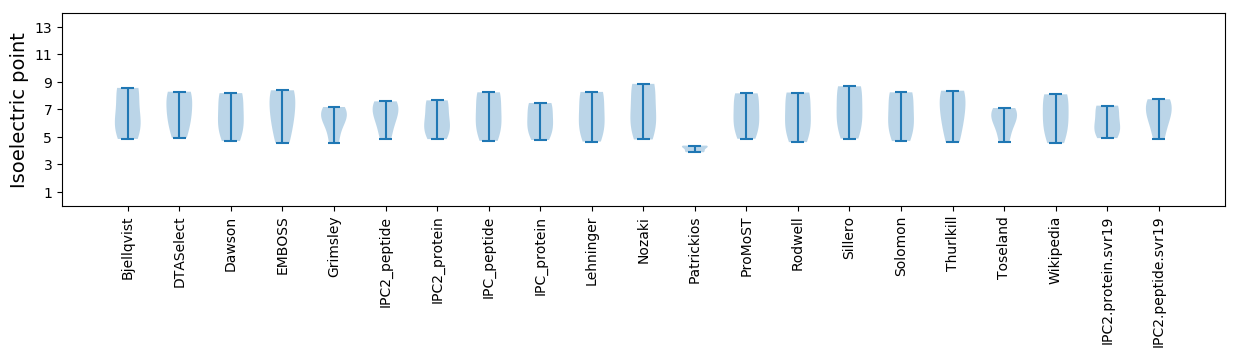
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1GMU0|A0A2P1GMU0_9RHAB Glycoprotein OS=Hainan black-spectacled toad dimarhabdovirus OX=2116358 PE=4 SV=1
MM1 pKa = 7.5NPLKK5 pKa = 10.54LFKK8 pKa = 10.35SGKK11 pKa = 9.42SEE13 pKa = 3.67VSPFINEE20 pKa = 3.85RR21 pKa = 11.84QPAEE25 pKa = 4.07NPYY28 pKa = 10.74GPTAPVEE35 pKa = 3.97TSSIYY40 pKa = 10.45KK41 pKa = 10.44LEE43 pKa = 4.07GKK45 pKa = 9.53ICLSLNKK52 pKa = 9.73PLEE55 pKa = 4.43DD56 pKa = 4.03PYY58 pKa = 11.85ALRR61 pKa = 11.84EE62 pKa = 3.97VLGLLEE68 pKa = 4.41KK69 pKa = 10.42NYY71 pKa = 11.11SGIHH75 pKa = 4.24QNKK78 pKa = 9.56YY79 pKa = 10.88LLLLAWNLTGCTLTRR94 pKa = 11.84GKK96 pKa = 9.95EE97 pKa = 3.97VAGVWTYY104 pKa = 7.52QTAIEE109 pKa = 4.33GFIDD113 pKa = 3.18IDD115 pKa = 4.08FNGEE119 pKa = 3.96TEE121 pKa = 4.3PCSEE125 pKa = 4.94ADD127 pKa = 3.87RR128 pKa = 11.84LWFEE132 pKa = 4.92SNVTGWTSGRR142 pKa = 11.84ICGEE146 pKa = 3.49AGFRR150 pKa = 11.84KK151 pKa = 9.89CAGCRR156 pKa = 11.84GEE158 pKa = 5.13SVTKK162 pKa = 10.72VLEE165 pKa = 3.97KK166 pKa = 10.22TGNKK170 pKa = 9.01LVPISQLLVDD180 pKa = 3.98SCIKK184 pKa = 10.55CRR186 pKa = 11.84VGTKK190 pKa = 10.3RR191 pKa = 11.84LVLYY195 pKa = 9.77KK196 pKa = 10.82AKK198 pKa = 10.73
MM1 pKa = 7.5NPLKK5 pKa = 10.54LFKK8 pKa = 10.35SGKK11 pKa = 9.42SEE13 pKa = 3.67VSPFINEE20 pKa = 3.85RR21 pKa = 11.84QPAEE25 pKa = 4.07NPYY28 pKa = 10.74GPTAPVEE35 pKa = 3.97TSSIYY40 pKa = 10.45KK41 pKa = 10.44LEE43 pKa = 4.07GKK45 pKa = 9.53ICLSLNKK52 pKa = 9.73PLEE55 pKa = 4.43DD56 pKa = 4.03PYY58 pKa = 11.85ALRR61 pKa = 11.84EE62 pKa = 3.97VLGLLEE68 pKa = 4.41KK69 pKa = 10.42NYY71 pKa = 11.11SGIHH75 pKa = 4.24QNKK78 pKa = 9.56YY79 pKa = 10.88LLLLAWNLTGCTLTRR94 pKa = 11.84GKK96 pKa = 9.95EE97 pKa = 3.97VAGVWTYY104 pKa = 7.52QTAIEE109 pKa = 4.33GFIDD113 pKa = 3.18IDD115 pKa = 4.08FNGEE119 pKa = 3.96TEE121 pKa = 4.3PCSEE125 pKa = 4.94ADD127 pKa = 3.87RR128 pKa = 11.84LWFEE132 pKa = 4.92SNVTGWTSGRR142 pKa = 11.84ICGEE146 pKa = 3.49AGFRR150 pKa = 11.84KK151 pKa = 9.89CAGCRR156 pKa = 11.84GEE158 pKa = 5.13SVTKK162 pKa = 10.72VLEE165 pKa = 3.97KK166 pKa = 10.22TGNKK170 pKa = 9.01LVPISQLLVDD180 pKa = 3.98SCIKK184 pKa = 10.55CRR186 pKa = 11.84VGTKK190 pKa = 10.3RR191 pKa = 11.84LVLYY195 pKa = 9.77KK196 pKa = 10.82AKK198 pKa = 10.73
Molecular weight: 21.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3510 |
198 |
2093 |
702.0 |
79.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.903 ± 0.884 | 2.194 ± 0.207 |
4.957 ± 0.433 | 7.236 ± 0.934 |
4.672 ± 0.366 | 6.866 ± 0.593 |
2.023 ± 0.284 | 7.009 ± 0.903 |
7.379 ± 0.552 | 9.459 ± 0.328 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.308 ± 0.263 | 4.986 ± 0.126 |
4.131 ± 0.575 | 2.764 ± 0.197 |
4.815 ± 0.345 | 8.006 ± 0.449 |
5.897 ± 0.337 | 5.869 ± 0.388 |
1.88 ± 0.112 | 3.647 ± 0.339 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
