
Scophthalmus maximus rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
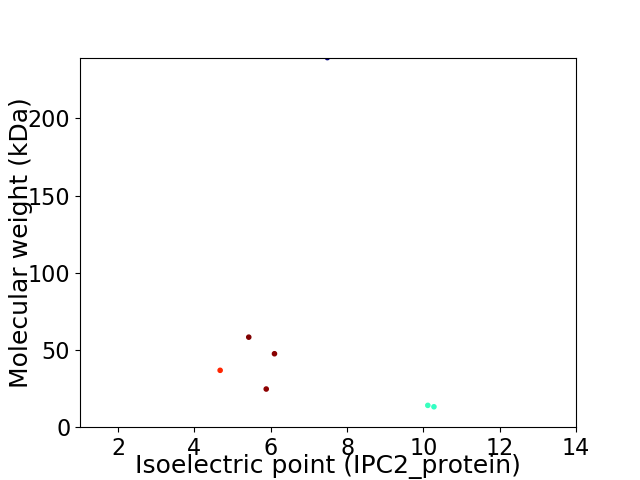
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
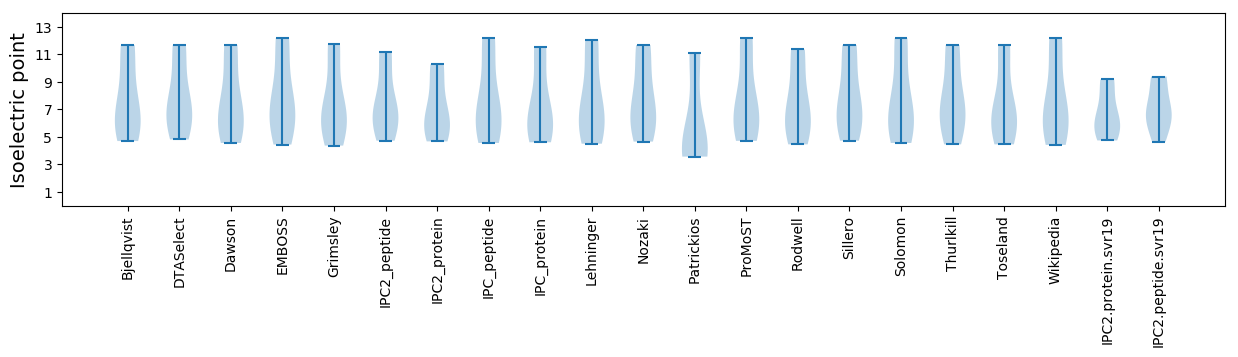
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E7D0U3|E7D0U3_9RHAB C protein OS=Scophthalmus maximus rhabdovirus OX=936149 GN=C PE=4 SV=1
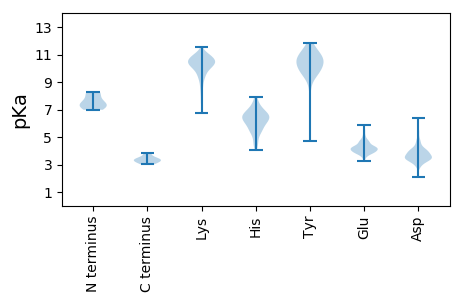
MM1 pKa = 7.29SHH3 pKa = 6.7GLSKK7 pKa = 10.88SRR9 pKa = 11.84TFEE12 pKa = 4.28EE13 pKa = 4.11ITDD16 pKa = 3.77VEE18 pKa = 4.29QVLDD22 pKa = 4.3NIRR25 pKa = 11.84QTEE28 pKa = 4.38SKK30 pKa = 10.51LGTSAGAGASDD41 pKa = 3.8EE42 pKa = 4.63PYY44 pKa = 10.15PAGGDD49 pKa = 3.62LPPASGGFRR58 pKa = 11.84YY59 pKa = 10.0LSSQEE64 pKa = 3.69EE65 pKa = 4.32GSYY68 pKa = 10.5KK69 pKa = 10.45EE70 pKa = 5.82EE71 pKa = 3.86EE72 pKa = 4.54DD73 pKa = 3.08NWLINQSRR81 pKa = 11.84AAKK84 pKa = 9.91LVAPLADD91 pKa = 3.52YY92 pKa = 10.87RR93 pKa = 11.84LAAAADD99 pKa = 4.1KK100 pKa = 10.98DD101 pKa = 4.03PSPEE105 pKa = 4.17PEE107 pKa = 3.86DD108 pKa = 3.22QRR110 pKa = 11.84RR111 pKa = 11.84EE112 pKa = 3.8IEE114 pKa = 3.92EE115 pKa = 4.64RR116 pKa = 11.84IAEE119 pKa = 4.21AHH121 pKa = 5.98DD122 pKa = 3.5QMLKK126 pKa = 10.82FEE128 pKa = 4.79MFPYY132 pKa = 9.84TYY134 pKa = 10.93FMPDD138 pKa = 3.23YY139 pKa = 10.37LKK141 pKa = 10.99SEE143 pKa = 4.26DD144 pKa = 3.72QQEE147 pKa = 4.25LVHH150 pKa = 6.81FLTEE154 pKa = 3.71ILGPRR159 pKa = 11.84NSNRR163 pKa = 11.84DD164 pKa = 2.94FWIRR168 pKa = 11.84GTHH171 pKa = 5.82IPTMCWRR178 pKa = 11.84NGDD181 pKa = 3.41MHH183 pKa = 7.82IQDD186 pKa = 4.4DD187 pKa = 4.58VEE189 pKa = 4.68SKK191 pKa = 10.89SSGVSGLTKK200 pKa = 10.37ALKK203 pKa = 10.05SVQFVDD209 pKa = 4.53PGEE212 pKa = 4.05QDD214 pKa = 3.24SATVEE219 pKa = 4.19SEE221 pKa = 3.42MDD223 pKa = 3.73RR224 pKa = 11.84DD225 pKa = 3.8LDD227 pKa = 4.04CYY229 pKa = 11.19SIGEE233 pKa = 4.19SSLVSEE239 pKa = 5.05FMLDD243 pKa = 3.22TSDD246 pKa = 5.62LMEE249 pKa = 4.49SLNRR253 pKa = 11.84EE254 pKa = 3.82WLVATKK260 pKa = 10.24KK261 pKa = 10.8GRR263 pKa = 11.84WKK265 pKa = 9.53TLGPLFLRR273 pKa = 11.84PLAQDD278 pKa = 3.26RR279 pKa = 11.84ALMKK283 pKa = 10.28TILSEE288 pKa = 3.98MRR290 pKa = 11.84EE291 pKa = 3.95KK292 pKa = 10.93DD293 pKa = 3.15VTEE296 pKa = 4.08QMSCLLTSNEE306 pKa = 3.8LTSRR310 pKa = 11.84FVRR313 pKa = 11.84TLDD316 pKa = 4.1LGTLKK321 pKa = 10.44MKK323 pKa = 10.85GSFF326 pKa = 3.44
MM1 pKa = 7.29SHH3 pKa = 6.7GLSKK7 pKa = 10.88SRR9 pKa = 11.84TFEE12 pKa = 4.28EE13 pKa = 4.11ITDD16 pKa = 3.77VEE18 pKa = 4.29QVLDD22 pKa = 4.3NIRR25 pKa = 11.84QTEE28 pKa = 4.38SKK30 pKa = 10.51LGTSAGAGASDD41 pKa = 3.8EE42 pKa = 4.63PYY44 pKa = 10.15PAGGDD49 pKa = 3.62LPPASGGFRR58 pKa = 11.84YY59 pKa = 10.0LSSQEE64 pKa = 3.69EE65 pKa = 4.32GSYY68 pKa = 10.5KK69 pKa = 10.45EE70 pKa = 5.82EE71 pKa = 3.86EE72 pKa = 4.54DD73 pKa = 3.08NWLINQSRR81 pKa = 11.84AAKK84 pKa = 9.91LVAPLADD91 pKa = 3.52YY92 pKa = 10.87RR93 pKa = 11.84LAAAADD99 pKa = 4.1KK100 pKa = 10.98DD101 pKa = 4.03PSPEE105 pKa = 4.17PEE107 pKa = 3.86DD108 pKa = 3.22QRR110 pKa = 11.84RR111 pKa = 11.84EE112 pKa = 3.8IEE114 pKa = 3.92EE115 pKa = 4.64RR116 pKa = 11.84IAEE119 pKa = 4.21AHH121 pKa = 5.98DD122 pKa = 3.5QMLKK126 pKa = 10.82FEE128 pKa = 4.79MFPYY132 pKa = 9.84TYY134 pKa = 10.93FMPDD138 pKa = 3.23YY139 pKa = 10.37LKK141 pKa = 10.99SEE143 pKa = 4.26DD144 pKa = 3.72QQEE147 pKa = 4.25LVHH150 pKa = 6.81FLTEE154 pKa = 3.71ILGPRR159 pKa = 11.84NSNRR163 pKa = 11.84DD164 pKa = 2.94FWIRR168 pKa = 11.84GTHH171 pKa = 5.82IPTMCWRR178 pKa = 11.84NGDD181 pKa = 3.41MHH183 pKa = 7.82IQDD186 pKa = 4.4DD187 pKa = 4.58VEE189 pKa = 4.68SKK191 pKa = 10.89SSGVSGLTKK200 pKa = 10.37ALKK203 pKa = 10.05SVQFVDD209 pKa = 4.53PGEE212 pKa = 4.05QDD214 pKa = 3.24SATVEE219 pKa = 4.19SEE221 pKa = 3.42MDD223 pKa = 3.73RR224 pKa = 11.84DD225 pKa = 3.8LDD227 pKa = 4.04CYY229 pKa = 11.19SIGEE233 pKa = 4.19SSLVSEE239 pKa = 5.05FMLDD243 pKa = 3.22TSDD246 pKa = 5.62LMEE249 pKa = 4.49SLNRR253 pKa = 11.84EE254 pKa = 3.82WLVATKK260 pKa = 10.24KK261 pKa = 10.8GRR263 pKa = 11.84WKK265 pKa = 9.53TLGPLFLRR273 pKa = 11.84PLAQDD278 pKa = 3.26RR279 pKa = 11.84ALMKK283 pKa = 10.28TILSEE288 pKa = 3.98MRR290 pKa = 11.84EE291 pKa = 3.95KK292 pKa = 10.93DD293 pKa = 3.15VTEE296 pKa = 4.08QMSCLLTSNEE306 pKa = 3.8LTSRR310 pKa = 11.84FVRR313 pKa = 11.84TLDD316 pKa = 4.1LGTLKK321 pKa = 10.44MKK323 pKa = 10.85GSFF326 pKa = 3.44
Molecular weight: 36.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E7D0U2|E7D0U2_9RHAB Phosphoprotein OS=Scophthalmus maximus rhabdovirus OX=936149 GN=P PE=4 SV=1
MM1 pKa = 6.98EE2 pKa = 4.44QVAKK6 pKa = 10.81CLMDD10 pKa = 3.82YY11 pKa = 10.33QNRR14 pKa = 11.84AHH16 pKa = 6.91LRR18 pKa = 11.84RR19 pKa = 11.84SRR21 pKa = 11.84MLNRR25 pKa = 11.84YY26 pKa = 4.69WTTYY30 pKa = 10.84DD31 pKa = 3.41KK32 pKa = 11.16QSPSWGRR39 pKa = 11.84LLEE42 pKa = 4.56RR43 pKa = 11.84EE44 pKa = 4.04RR45 pKa = 11.84QMNPIPPEE53 pKa = 4.16VIYY56 pKa = 10.37HH57 pKa = 5.31QLPVDD62 pKa = 5.33LGTCPLKK69 pKa = 10.69RR70 pKa = 11.84RR71 pKa = 11.84GAIRR75 pKa = 11.84RR76 pKa = 11.84KK77 pKa = 8.7RR78 pKa = 11.84TTGSSTNPEE87 pKa = 3.99PQSWWLRR94 pKa = 11.84LPIIGWLLQRR104 pKa = 11.84IRR106 pKa = 11.84TLVLNRR112 pKa = 11.84RR113 pKa = 11.84TNGGRR118 pKa = 3.21
MM1 pKa = 6.98EE2 pKa = 4.44QVAKK6 pKa = 10.81CLMDD10 pKa = 3.82YY11 pKa = 10.33QNRR14 pKa = 11.84AHH16 pKa = 6.91LRR18 pKa = 11.84RR19 pKa = 11.84SRR21 pKa = 11.84MLNRR25 pKa = 11.84YY26 pKa = 4.69WTTYY30 pKa = 10.84DD31 pKa = 3.41KK32 pKa = 11.16QSPSWGRR39 pKa = 11.84LLEE42 pKa = 4.56RR43 pKa = 11.84EE44 pKa = 4.04RR45 pKa = 11.84QMNPIPPEE53 pKa = 4.16VIYY56 pKa = 10.37HH57 pKa = 5.31QLPVDD62 pKa = 5.33LGTCPLKK69 pKa = 10.69RR70 pKa = 11.84RR71 pKa = 11.84GAIRR75 pKa = 11.84RR76 pKa = 11.84KK77 pKa = 8.7RR78 pKa = 11.84TTGSSTNPEE87 pKa = 3.99PQSWWLRR94 pKa = 11.84LPIIGWLLQRR104 pKa = 11.84IRR106 pKa = 11.84TLVLNRR112 pKa = 11.84RR113 pKa = 11.84TNGGRR118 pKa = 3.21
Molecular weight: 14.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3843 |
110 |
2116 |
549.0 |
62.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.557 ± 1.05 | 1.717 ± 0.353 |
5.491 ± 0.418 | 6.193 ± 0.548 |
3.435 ± 0.391 | 6.063 ± 0.291 |
2.42 ± 0.281 | 5.517 ± 0.564 |
5.673 ± 0.344 | 9.576 ± 0.701 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.227 ± 0.119 | 4.059 ± 0.266 |
4.97 ± 0.434 | 3.487 ± 0.208 |
6.089 ± 0.863 | 8.171 ± 0.722 |
6.167 ± 0.279 | 5.881 ± 0.746 |
2.212 ± 0.202 | 3.097 ± 0.309 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
