
Hermit crab associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.09
Get precalculated fractions of proteins
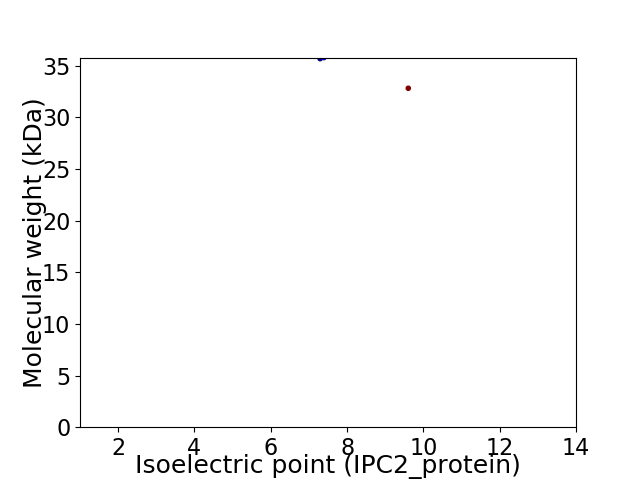
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
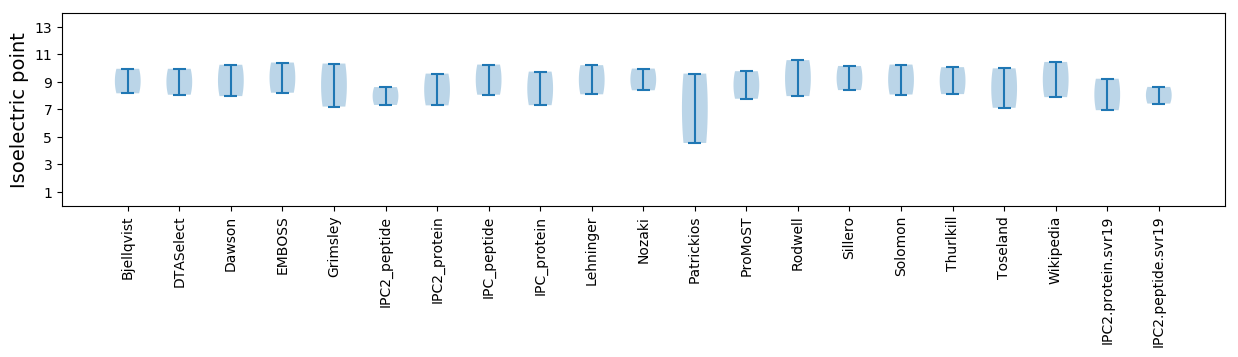
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RLN6|A0A0K1RLN6_9CIRC Putative capsid protein OS=Hermit crab associated circular virus OX=1692252 PE=4 SV=1
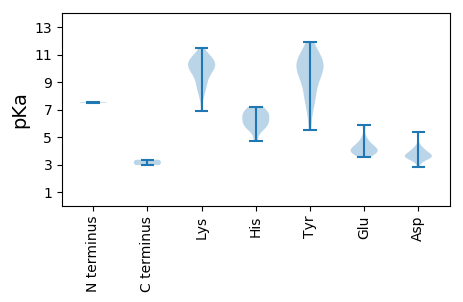
MM1 pKa = 7.47PRR3 pKa = 11.84DD4 pKa = 3.53APAKK8 pKa = 9.06QWCFTLNNYY17 pKa = 7.88TPAEE21 pKa = 3.96LTAIVDD27 pKa = 3.68SAGNFDD33 pKa = 4.15YY34 pKa = 11.33LCFGRR39 pKa = 11.84EE40 pKa = 3.57RR41 pKa = 11.84GNNNTPHH48 pKa = 5.96LQGYY52 pKa = 9.48LILKK56 pKa = 6.87EE57 pKa = 4.27KK58 pKa = 10.76KK59 pKa = 7.99RR60 pKa = 11.84FSYY63 pKa = 10.31VRR65 pKa = 11.84QLAGLEE71 pKa = 3.88RR72 pKa = 11.84AHH74 pKa = 6.68WEE76 pKa = 4.15KK77 pKa = 10.75KK78 pKa = 9.95SPRR81 pKa = 11.84STPKK85 pKa = 10.09QASDD89 pKa = 3.59YY90 pKa = 10.18CKK92 pKa = 10.48KK93 pKa = 10.86DD94 pKa = 2.83GDD96 pKa = 3.73YY97 pKa = 11.43DD98 pKa = 3.93EE99 pKa = 5.87YY100 pKa = 11.9GEE102 pKa = 5.69LPTKK106 pKa = 10.56KK107 pKa = 9.78QGQRR111 pKa = 11.84TDD113 pKa = 3.64FDD115 pKa = 4.24EE116 pKa = 4.57LKK118 pKa = 10.64EE119 pKa = 4.19WIKK122 pKa = 10.72EE123 pKa = 3.84QDD125 pKa = 3.06HH126 pKa = 6.62RR127 pKa = 11.84PTDD130 pKa = 3.47RR131 pKa = 11.84EE132 pKa = 3.97VAEE135 pKa = 4.78EE136 pKa = 5.06FPSLWGRR143 pKa = 11.84YY144 pKa = 7.82RR145 pKa = 11.84SACISFLDD153 pKa = 4.41LFSPHH158 pKa = 6.48PTLVQGTLRR167 pKa = 11.84PWQEE171 pKa = 4.13DD172 pKa = 3.89LNTRR176 pKa = 11.84LSAPPNDD183 pKa = 3.91RR184 pKa = 11.84DD185 pKa = 3.54VMFVVDD191 pKa = 4.08EE192 pKa = 4.62NGNSGKK198 pKa = 10.12SWFIRR203 pKa = 11.84YY204 pKa = 9.39LMTEE208 pKa = 4.17RR209 pKa = 11.84PDD211 pKa = 3.62DD212 pKa = 3.83VQMLSIGKK220 pKa = 9.53RR221 pKa = 11.84DD222 pKa = 4.17DD223 pKa = 3.49LAHH226 pKa = 7.2AIDD229 pKa = 3.96PAKK232 pKa = 10.57KK233 pKa = 9.81IFFFDD238 pKa = 3.55VPRR241 pKa = 11.84GGMEE245 pKa = 3.96FMQYY249 pKa = 11.06AVLEE253 pKa = 4.12QLKK256 pKa = 9.96NRR258 pKa = 11.84LVFSPKK264 pKa = 9.36YY265 pKa = 7.58EE266 pKa = 3.98SRR268 pKa = 11.84MKK270 pKa = 10.2VLHH273 pKa = 6.83HH274 pKa = 6.76IPHH277 pKa = 5.61VVVFCNEE284 pKa = 3.68EE285 pKa = 3.72PDD287 pKa = 3.52RR288 pKa = 11.84TKK290 pKa = 11.02LSRR293 pKa = 11.84DD294 pKa = 3.3RR295 pKa = 11.84FRR297 pKa = 11.84VTHH300 pKa = 5.94IRR302 pKa = 11.84QVV304 pKa = 2.96
MM1 pKa = 7.47PRR3 pKa = 11.84DD4 pKa = 3.53APAKK8 pKa = 9.06QWCFTLNNYY17 pKa = 7.88TPAEE21 pKa = 3.96LTAIVDD27 pKa = 3.68SAGNFDD33 pKa = 4.15YY34 pKa = 11.33LCFGRR39 pKa = 11.84EE40 pKa = 3.57RR41 pKa = 11.84GNNNTPHH48 pKa = 5.96LQGYY52 pKa = 9.48LILKK56 pKa = 6.87EE57 pKa = 4.27KK58 pKa = 10.76KK59 pKa = 7.99RR60 pKa = 11.84FSYY63 pKa = 10.31VRR65 pKa = 11.84QLAGLEE71 pKa = 3.88RR72 pKa = 11.84AHH74 pKa = 6.68WEE76 pKa = 4.15KK77 pKa = 10.75KK78 pKa = 9.95SPRR81 pKa = 11.84STPKK85 pKa = 10.09QASDD89 pKa = 3.59YY90 pKa = 10.18CKK92 pKa = 10.48KK93 pKa = 10.86DD94 pKa = 2.83GDD96 pKa = 3.73YY97 pKa = 11.43DD98 pKa = 3.93EE99 pKa = 5.87YY100 pKa = 11.9GEE102 pKa = 5.69LPTKK106 pKa = 10.56KK107 pKa = 9.78QGQRR111 pKa = 11.84TDD113 pKa = 3.64FDD115 pKa = 4.24EE116 pKa = 4.57LKK118 pKa = 10.64EE119 pKa = 4.19WIKK122 pKa = 10.72EE123 pKa = 3.84QDD125 pKa = 3.06HH126 pKa = 6.62RR127 pKa = 11.84PTDD130 pKa = 3.47RR131 pKa = 11.84EE132 pKa = 3.97VAEE135 pKa = 4.78EE136 pKa = 5.06FPSLWGRR143 pKa = 11.84YY144 pKa = 7.82RR145 pKa = 11.84SACISFLDD153 pKa = 4.41LFSPHH158 pKa = 6.48PTLVQGTLRR167 pKa = 11.84PWQEE171 pKa = 4.13DD172 pKa = 3.89LNTRR176 pKa = 11.84LSAPPNDD183 pKa = 3.91RR184 pKa = 11.84DD185 pKa = 3.54VMFVVDD191 pKa = 4.08EE192 pKa = 4.62NGNSGKK198 pKa = 10.12SWFIRR203 pKa = 11.84YY204 pKa = 9.39LMTEE208 pKa = 4.17RR209 pKa = 11.84PDD211 pKa = 3.62DD212 pKa = 3.83VQMLSIGKK220 pKa = 9.53RR221 pKa = 11.84DD222 pKa = 4.17DD223 pKa = 3.49LAHH226 pKa = 7.2AIDD229 pKa = 3.96PAKK232 pKa = 10.57KK233 pKa = 9.81IFFFDD238 pKa = 3.55VPRR241 pKa = 11.84GGMEE245 pKa = 3.96FMQYY249 pKa = 11.06AVLEE253 pKa = 4.12QLKK256 pKa = 9.96NRR258 pKa = 11.84LVFSPKK264 pKa = 9.36YY265 pKa = 7.58EE266 pKa = 3.98SRR268 pKa = 11.84MKK270 pKa = 10.2VLHH273 pKa = 6.83HH274 pKa = 6.76IPHH277 pKa = 5.61VVVFCNEE284 pKa = 3.68EE285 pKa = 3.72PDD287 pKa = 3.52RR288 pKa = 11.84TKK290 pKa = 11.02LSRR293 pKa = 11.84DD294 pKa = 3.3RR295 pKa = 11.84FRR297 pKa = 11.84VTHH300 pKa = 5.94IRR302 pKa = 11.84QVV304 pKa = 2.96
Molecular weight: 35.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RLN6|A0A0K1RLN6_9CIRC Putative capsid protein OS=Hermit crab associated circular virus OX=1692252 PE=4 SV=1
MM1 pKa = 7.53TKK3 pKa = 10.32FLTPPRR9 pKa = 11.84PSSKK13 pKa = 9.97RR14 pKa = 11.84LRR16 pKa = 11.84AVSPKK21 pKa = 8.94MQTGTPEE28 pKa = 4.01PAHH31 pKa = 6.23TIMPRR36 pKa = 11.84GISTHH41 pKa = 4.7MRR43 pKa = 11.84KK44 pKa = 9.09SAYY47 pKa = 9.37HH48 pKa = 6.23RR49 pKa = 11.84NLGRR53 pKa = 11.84RR54 pKa = 11.84VGKK57 pKa = 10.34YY58 pKa = 5.52STRR61 pKa = 11.84RR62 pKa = 11.84TLYY65 pKa = 10.05TGFKK69 pKa = 10.17NDD71 pKa = 3.7LVSKK75 pKa = 8.46KK76 pKa = 8.65TYY78 pKa = 7.5WSRR81 pKa = 11.84LIRR84 pKa = 11.84IPHH87 pKa = 5.69YY88 pKa = 10.37GAGQEE93 pKa = 3.84EE94 pKa = 4.81HH95 pKa = 5.74IKK97 pKa = 10.02YY98 pKa = 10.27RR99 pKa = 11.84RR100 pKa = 11.84GAYY103 pKa = 10.2CNVRR107 pKa = 11.84GVKK110 pKa = 9.53INYY113 pKa = 6.65WFSMDD118 pKa = 5.37ANTQYY123 pKa = 10.77PVQVRR128 pKa = 11.84WAIINPKK135 pKa = 9.52VNEE138 pKa = 4.31GTDD141 pKa = 3.57LTLVTNPEE149 pKa = 3.73NFFISKK155 pKa = 10.57DD156 pKa = 3.38PTTEE160 pKa = 3.61QSEE163 pKa = 4.39NFPTTGNYY171 pKa = 8.73FKK173 pKa = 11.5YY174 pKa = 9.78MNRR177 pKa = 11.84KK178 pKa = 8.68INRR181 pKa = 11.84EE182 pKa = 3.72LYY184 pKa = 10.13GVLKK188 pKa = 10.58EE189 pKa = 4.41GTFTMSIDD197 pKa = 3.49PAAAVPNPGVVYY209 pKa = 10.55RR210 pKa = 11.84PGSAVKK216 pKa = 10.24KK217 pKa = 8.52IALYY221 pKa = 10.77VPVNRR226 pKa = 11.84QMKK229 pKa = 7.92WNNVVAGDD237 pKa = 3.8ANGYY241 pKa = 9.0PEE243 pKa = 4.03QNLYY247 pKa = 9.27FVWWYY252 pKa = 8.42DD253 pKa = 3.38TAANDD258 pKa = 3.63NTNPSASPLVQEE270 pKa = 4.48FHH272 pKa = 7.1EE273 pKa = 4.15KK274 pKa = 9.9TIYY277 pKa = 10.33FKK279 pKa = 10.97NSQMFAA285 pKa = 3.35
MM1 pKa = 7.53TKK3 pKa = 10.32FLTPPRR9 pKa = 11.84PSSKK13 pKa = 9.97RR14 pKa = 11.84LRR16 pKa = 11.84AVSPKK21 pKa = 8.94MQTGTPEE28 pKa = 4.01PAHH31 pKa = 6.23TIMPRR36 pKa = 11.84GISTHH41 pKa = 4.7MRR43 pKa = 11.84KK44 pKa = 9.09SAYY47 pKa = 9.37HH48 pKa = 6.23RR49 pKa = 11.84NLGRR53 pKa = 11.84RR54 pKa = 11.84VGKK57 pKa = 10.34YY58 pKa = 5.52STRR61 pKa = 11.84RR62 pKa = 11.84TLYY65 pKa = 10.05TGFKK69 pKa = 10.17NDD71 pKa = 3.7LVSKK75 pKa = 8.46KK76 pKa = 8.65TYY78 pKa = 7.5WSRR81 pKa = 11.84LIRR84 pKa = 11.84IPHH87 pKa = 5.69YY88 pKa = 10.37GAGQEE93 pKa = 3.84EE94 pKa = 4.81HH95 pKa = 5.74IKK97 pKa = 10.02YY98 pKa = 10.27RR99 pKa = 11.84RR100 pKa = 11.84GAYY103 pKa = 10.2CNVRR107 pKa = 11.84GVKK110 pKa = 9.53INYY113 pKa = 6.65WFSMDD118 pKa = 5.37ANTQYY123 pKa = 10.77PVQVRR128 pKa = 11.84WAIINPKK135 pKa = 9.52VNEE138 pKa = 4.31GTDD141 pKa = 3.57LTLVTNPEE149 pKa = 3.73NFFISKK155 pKa = 10.57DD156 pKa = 3.38PTTEE160 pKa = 3.61QSEE163 pKa = 4.39NFPTTGNYY171 pKa = 8.73FKK173 pKa = 11.5YY174 pKa = 9.78MNRR177 pKa = 11.84KK178 pKa = 8.68INRR181 pKa = 11.84EE182 pKa = 3.72LYY184 pKa = 10.13GVLKK188 pKa = 10.58EE189 pKa = 4.41GTFTMSIDD197 pKa = 3.49PAAAVPNPGVVYY209 pKa = 10.55RR210 pKa = 11.84PGSAVKK216 pKa = 10.24KK217 pKa = 8.52IALYY221 pKa = 10.77VPVNRR226 pKa = 11.84QMKK229 pKa = 7.92WNNVVAGDD237 pKa = 3.8ANGYY241 pKa = 9.0PEE243 pKa = 4.03QNLYY247 pKa = 9.27FVWWYY252 pKa = 8.42DD253 pKa = 3.38TAANDD258 pKa = 3.63NTNPSASPLVQEE270 pKa = 4.48FHH272 pKa = 7.1EE273 pKa = 4.15KK274 pKa = 9.9TIYY277 pKa = 10.33FKK279 pKa = 10.97NSQMFAA285 pKa = 3.35
Molecular weight: 32.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
589 |
285 |
304 |
294.5 |
34.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.603 ± 0.483 | 1.019 ± 0.453 |
5.433 ± 1.78 | 5.603 ± 0.944 |
4.924 ± 0.483 | 5.433 ± 0.361 |
2.547 ± 0.299 | 3.905 ± 0.445 |
6.961 ± 0.038 | 6.452 ± 1.282 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.716 ± 0.299 | 5.942 ± 1.443 |
6.961 ± 0.276 | 3.735 ± 0.391 |
7.81 ± 0.537 | 5.433 ± 0.123 |
6.282 ± 1.213 | 6.282 ± 0.499 |
2.037 ± 0.046 | 4.924 ± 0.944 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
