
Ostreid herpesvirus 1 (isolate France) (OsHV-1) (Pacific oyster herpesvirus)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Malacoherpesviridae; Ostreavirus; Ostreid herpesvirus 1
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
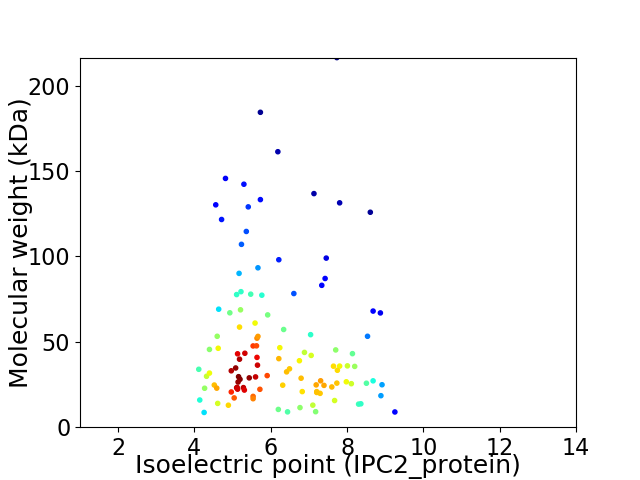
Virtual 2D-PAGE plot for 116 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
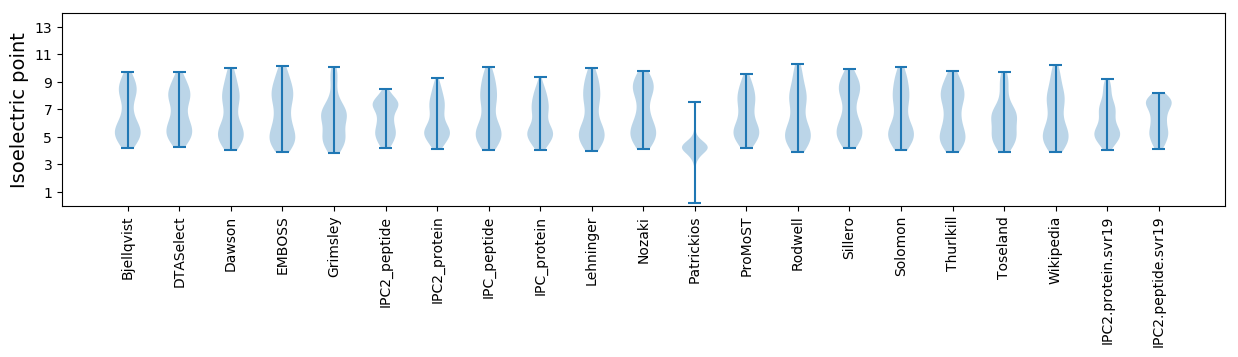
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q6R797|Y118_OSHVF Putative RING finger protein ORF118 OS=Ostreid herpesvirus 1 (isolate France) OX=654903 GN=ORF118 PE=4 SV=1
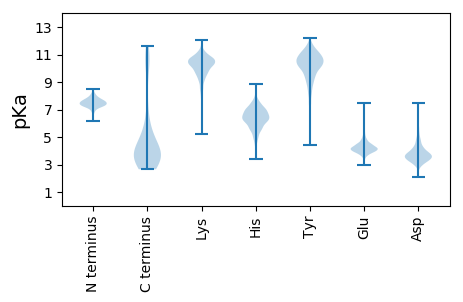
MM1 pKa = 7.35AAVALTYY8 pKa = 7.45MIPPATPLAIIDD20 pKa = 4.04FDD22 pKa = 4.19MLPEE26 pKa = 4.99DD27 pKa = 5.0DD28 pKa = 4.37EE29 pKa = 5.07SEE31 pKa = 4.37EE32 pKa = 4.29VGFEE36 pKa = 4.44YY37 pKa = 10.77EE38 pKa = 6.37DD39 pKa = 4.09LLDD42 pKa = 3.82WQLKK46 pKa = 8.26EE47 pKa = 4.1AVDD50 pKa = 3.96RR51 pKa = 11.84RR52 pKa = 11.84DD53 pKa = 3.65LEE55 pKa = 4.25SFTILIEE62 pKa = 4.31EE63 pKa = 4.36NWCHH67 pKa = 7.38DD68 pKa = 3.72LTLEE72 pKa = 5.16NGDD75 pKa = 3.76NLLGYY80 pKa = 8.69MIKK83 pKa = 10.0QGHH86 pKa = 6.78ALTEE90 pKa = 3.74EE91 pKa = 4.44WYY93 pKa = 10.37DD94 pKa = 4.08LIGVTTTCHH103 pKa = 6.3ANPEE107 pKa = 4.24GEE109 pKa = 4.43LPIDD113 pKa = 4.48LLRR116 pKa = 11.84KK117 pKa = 9.47HH118 pKa = 6.55LGSCRR123 pKa = 11.84CNLEE127 pKa = 4.43SMNCDD132 pKa = 2.76LVEE135 pKa = 4.04YY136 pKa = 10.26IIRR139 pKa = 11.84TCYY142 pKa = 9.94AHH144 pKa = 7.46PINRR148 pKa = 11.84DD149 pKa = 3.45YY150 pKa = 11.26VYY152 pKa = 11.11HH153 pKa = 7.39RR154 pKa = 11.84MDD156 pKa = 3.44TSRR159 pKa = 11.84YY160 pKa = 4.41THH162 pKa = 6.42KK163 pKa = 10.76RR164 pKa = 11.84LNRR167 pKa = 11.84NHH169 pKa = 6.65NLTLSIEE176 pKa = 4.13KK177 pKa = 10.48KK178 pKa = 9.98PIIVEE183 pKa = 4.06APPMEE188 pKa = 4.35EE189 pKa = 4.02EE190 pKa = 4.55EE191 pKa = 4.07ISEE194 pKa = 4.57VEE196 pKa = 3.99DD197 pKa = 3.71ALNVLQRR204 pKa = 11.84LCAQEE209 pKa = 4.77EE210 pKa = 4.61GDD212 pKa = 3.77NKK214 pKa = 10.5EE215 pKa = 5.05AEE217 pKa = 4.58TNNNNYY223 pKa = 8.53VFPWMTPAYY232 pKa = 7.88EE233 pKa = 4.46LPEE236 pKa = 3.88QLTFYY241 pKa = 10.57DD242 pKa = 3.65MPMTPFNCNNVMYY255 pKa = 10.41CC256 pKa = 4.09
MM1 pKa = 7.35AAVALTYY8 pKa = 7.45MIPPATPLAIIDD20 pKa = 4.04FDD22 pKa = 4.19MLPEE26 pKa = 4.99DD27 pKa = 5.0DD28 pKa = 4.37EE29 pKa = 5.07SEE31 pKa = 4.37EE32 pKa = 4.29VGFEE36 pKa = 4.44YY37 pKa = 10.77EE38 pKa = 6.37DD39 pKa = 4.09LLDD42 pKa = 3.82WQLKK46 pKa = 8.26EE47 pKa = 4.1AVDD50 pKa = 3.96RR51 pKa = 11.84RR52 pKa = 11.84DD53 pKa = 3.65LEE55 pKa = 4.25SFTILIEE62 pKa = 4.31EE63 pKa = 4.36NWCHH67 pKa = 7.38DD68 pKa = 3.72LTLEE72 pKa = 5.16NGDD75 pKa = 3.76NLLGYY80 pKa = 8.69MIKK83 pKa = 10.0QGHH86 pKa = 6.78ALTEE90 pKa = 3.74EE91 pKa = 4.44WYY93 pKa = 10.37DD94 pKa = 4.08LIGVTTTCHH103 pKa = 6.3ANPEE107 pKa = 4.24GEE109 pKa = 4.43LPIDD113 pKa = 4.48LLRR116 pKa = 11.84KK117 pKa = 9.47HH118 pKa = 6.55LGSCRR123 pKa = 11.84CNLEE127 pKa = 4.43SMNCDD132 pKa = 2.76LVEE135 pKa = 4.04YY136 pKa = 10.26IIRR139 pKa = 11.84TCYY142 pKa = 9.94AHH144 pKa = 7.46PINRR148 pKa = 11.84DD149 pKa = 3.45YY150 pKa = 11.26VYY152 pKa = 11.11HH153 pKa = 7.39RR154 pKa = 11.84MDD156 pKa = 3.44TSRR159 pKa = 11.84YY160 pKa = 4.41THH162 pKa = 6.42KK163 pKa = 10.76RR164 pKa = 11.84LNRR167 pKa = 11.84NHH169 pKa = 6.65NLTLSIEE176 pKa = 4.13KK177 pKa = 10.48KK178 pKa = 9.98PIIVEE183 pKa = 4.06APPMEE188 pKa = 4.35EE189 pKa = 4.02EE190 pKa = 4.55EE191 pKa = 4.07ISEE194 pKa = 4.57VEE196 pKa = 3.99DD197 pKa = 3.71ALNVLQRR204 pKa = 11.84LCAQEE209 pKa = 4.77EE210 pKa = 4.61GDD212 pKa = 3.77NKK214 pKa = 10.5EE215 pKa = 5.05AEE217 pKa = 4.58TNNNNYY223 pKa = 8.53VFPWMTPAYY232 pKa = 7.88EE233 pKa = 4.46LPEE236 pKa = 3.88QLTFYY241 pKa = 10.57DD242 pKa = 3.65MPMTPFNCNNVMYY255 pKa = 10.41CC256 pKa = 4.09
Molecular weight: 29.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q6R7J0|Y034_OSHVF Uncharacterized protein ORF34 OS=Ostreid herpesvirus 1 (isolate France) OX=654903 GN=ORF34 PE=4 SV=1
MM1 pKa = 7.19STTEE5 pKa = 3.71QTVCEE10 pKa = 4.14IEE12 pKa = 4.23QEE14 pKa = 4.41SEE16 pKa = 4.53LIPAKK21 pKa = 9.63PQYY24 pKa = 10.65IIVKK28 pKa = 9.46KK29 pKa = 9.85PKK31 pKa = 7.87RR32 pKa = 11.84QAWQRR37 pKa = 11.84VLLLFRR43 pKa = 11.84IINMIVIWAALIALFVKK60 pKa = 10.24LYY62 pKa = 9.87ILRR65 pKa = 11.84GPIPRR70 pKa = 11.84SYY72 pKa = 10.89FHH74 pKa = 6.6YY75 pKa = 10.81
MM1 pKa = 7.19STTEE5 pKa = 3.71QTVCEE10 pKa = 4.14IEE12 pKa = 4.23QEE14 pKa = 4.41SEE16 pKa = 4.53LIPAKK21 pKa = 9.63PQYY24 pKa = 10.65IIVKK28 pKa = 9.46KK29 pKa = 9.85PKK31 pKa = 7.87RR32 pKa = 11.84QAWQRR37 pKa = 11.84VLLLFRR43 pKa = 11.84IINMIVIWAALIALFVKK60 pKa = 10.24LYY62 pKa = 9.87ILRR65 pKa = 11.84GPIPRR70 pKa = 11.84SYY72 pKa = 10.89FHH74 pKa = 6.6YY75 pKa = 10.81
Molecular weight: 8.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
51428 |
75 |
1878 |
443.3 |
50.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.095 ± 0.142 | 1.923 ± 0.122 |
6.85 ± 0.135 | 7.539 ± 0.175 |
4.122 ± 0.085 | 4.974 ± 0.121 |
1.925 ± 0.08 | 7.123 ± 0.154 |
7.982 ± 0.206 | 7.966 ± 0.166 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.535 ± 0.123 | 5.658 ± 0.128 |
4.305 ± 0.214 | 2.641 ± 0.071 |
4.964 ± 0.146 | 6.234 ± 0.143 |
6.213 ± 0.166 | 6.098 ± 0.104 |
0.842 ± 0.047 | 4.011 ± 0.1 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
