
Lewinella agarilytica
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Saprospiria; Saprospirales; Lewinellaceae; Lewinella
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
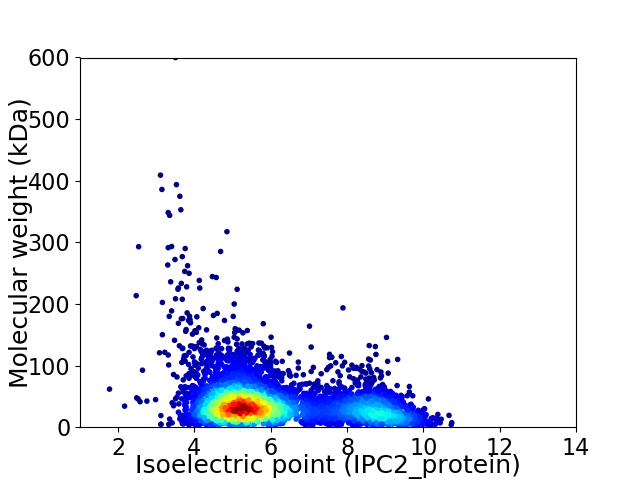
Virtual 2D-PAGE plot for 4889 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9J3Z8|A0A1H9J3Z8_9BACT Uncharacterized protein OS=Lewinella agarilytica OX=478744 GN=SAMN05444359_11678 PE=4 SV=1
MM1 pKa = 7.41LKK3 pKa = 10.39KK4 pKa = 10.77LSFLLAACLIVMTGCKK20 pKa = 9.71DD21 pKa = 3.54DD22 pKa = 4.09EE23 pKa = 4.47LAPVIIFNEE32 pKa = 4.29LEE34 pKa = 3.89VGAFPFLNEE43 pKa = 4.11LKK45 pKa = 10.12TGEE48 pKa = 4.43FDD50 pKa = 4.4LADD53 pKa = 4.05LAGSAYY59 pKa = 10.15EE60 pKa = 3.79MDD62 pKa = 4.08VYY64 pKa = 10.93FVDD67 pKa = 4.6GQAGANIAQYY77 pKa = 10.42NVYY80 pKa = 8.66VTFEE84 pKa = 4.93DD85 pKa = 3.97NNPSNGDD92 pKa = 3.34EE93 pKa = 4.28SSTAEE98 pKa = 3.92ALFRR102 pKa = 11.84SYY104 pKa = 11.21VPSDD108 pKa = 3.5FEE110 pKa = 4.75TIGDD114 pKa = 3.98KK115 pKa = 11.46GNLGTNVVIPFLDD128 pKa = 3.53AATAAGVDD136 pKa = 3.55ADD138 pKa = 4.28NVLSGDD144 pKa = 3.41RR145 pKa = 11.84FQFRR149 pKa = 11.84TEE151 pKa = 3.62IVTTTGTTFNSVNSTPAVTNAFGGLWNFNVTATCPLADD189 pKa = 3.76DD190 pKa = 4.5VFVGSYY196 pKa = 9.35NVSYY200 pKa = 10.67GYY202 pKa = 10.19IYY204 pKa = 10.65DD205 pKa = 3.63PFVIFGRR212 pKa = 11.84TIQGWLPAPMDD223 pKa = 3.58QNVTLEE229 pKa = 4.3LKK231 pKa = 10.69AGSTTVRR238 pKa = 11.84SFGTQYY244 pKa = 11.16LVPSDD249 pKa = 4.03LTSDD253 pKa = 3.14ITIDD257 pKa = 3.47LTFSCDD263 pKa = 2.92VVTAVSSDD271 pKa = 3.17SGVRR275 pKa = 11.84CSGAGWRR282 pKa = 11.84PIQINTASFDD292 pKa = 4.32LNDD295 pKa = 4.57DD296 pKa = 3.62STFTIEE302 pKa = 4.05FRR304 pKa = 11.84DD305 pKa = 3.48WGSDD309 pKa = 3.5EE310 pKa = 5.22NNGGCGNIAAQEE322 pKa = 4.05YY323 pKa = 9.9SVVFVKK329 pKa = 10.35QQ330 pKa = 3.04
MM1 pKa = 7.41LKK3 pKa = 10.39KK4 pKa = 10.77LSFLLAACLIVMTGCKK20 pKa = 9.71DD21 pKa = 3.54DD22 pKa = 4.09EE23 pKa = 4.47LAPVIIFNEE32 pKa = 4.29LEE34 pKa = 3.89VGAFPFLNEE43 pKa = 4.11LKK45 pKa = 10.12TGEE48 pKa = 4.43FDD50 pKa = 4.4LADD53 pKa = 4.05LAGSAYY59 pKa = 10.15EE60 pKa = 3.79MDD62 pKa = 4.08VYY64 pKa = 10.93FVDD67 pKa = 4.6GQAGANIAQYY77 pKa = 10.42NVYY80 pKa = 8.66VTFEE84 pKa = 4.93DD85 pKa = 3.97NNPSNGDD92 pKa = 3.34EE93 pKa = 4.28SSTAEE98 pKa = 3.92ALFRR102 pKa = 11.84SYY104 pKa = 11.21VPSDD108 pKa = 3.5FEE110 pKa = 4.75TIGDD114 pKa = 3.98KK115 pKa = 11.46GNLGTNVVIPFLDD128 pKa = 3.53AATAAGVDD136 pKa = 3.55ADD138 pKa = 4.28NVLSGDD144 pKa = 3.41RR145 pKa = 11.84FQFRR149 pKa = 11.84TEE151 pKa = 3.62IVTTTGTTFNSVNSTPAVTNAFGGLWNFNVTATCPLADD189 pKa = 3.76DD190 pKa = 4.5VFVGSYY196 pKa = 9.35NVSYY200 pKa = 10.67GYY202 pKa = 10.19IYY204 pKa = 10.65DD205 pKa = 3.63PFVIFGRR212 pKa = 11.84TIQGWLPAPMDD223 pKa = 3.58QNVTLEE229 pKa = 4.3LKK231 pKa = 10.69AGSTTVRR238 pKa = 11.84SFGTQYY244 pKa = 11.16LVPSDD249 pKa = 4.03LTSDD253 pKa = 3.14ITIDD257 pKa = 3.47LTFSCDD263 pKa = 2.92VVTAVSSDD271 pKa = 3.17SGVRR275 pKa = 11.84CSGAGWRR282 pKa = 11.84PIQINTASFDD292 pKa = 4.32LNDD295 pKa = 4.57DD296 pKa = 3.62STFTIEE302 pKa = 4.05FRR304 pKa = 11.84DD305 pKa = 3.48WGSDD309 pKa = 3.5EE310 pKa = 5.22NNGGCGNIAAQEE322 pKa = 4.05YY323 pKa = 9.9SVVFVKK329 pKa = 10.35QQ330 pKa = 3.04
Molecular weight: 35.57 kDa
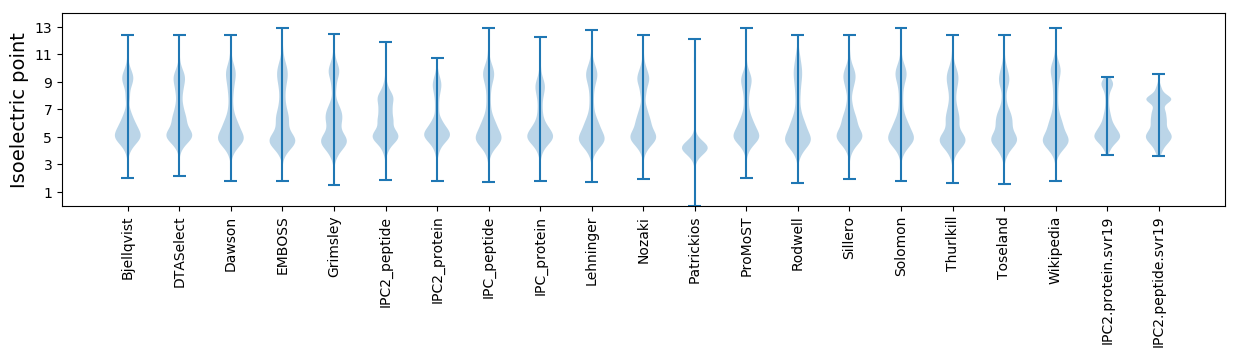
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9KXV2|A0A1H9KXV2_9BACT Lipoprotein-releasing system permease protein OS=Lewinella agarilytica OX=478744 GN=SAMN05444359_12234 PE=4 SV=1
MM1 pKa = 7.22GRR3 pKa = 11.84SPQVQGGLKK12 pKa = 9.26GQWGSLSLLPLCCSTVACTCVSAATFFWSNFSRR45 pKa = 11.84TCTSSPAGVKK55 pKa = 10.23RR56 pKa = 11.84ILVQYY61 pKa = 10.1NISLHH66 pKa = 6.33LFVLRR71 pKa = 11.84PSAYY75 pKa = 9.57FSAGFSFVEE84 pKa = 4.23ARR86 pKa = 11.84SKK88 pKa = 10.35FARR91 pKa = 11.84ARR93 pKa = 11.84AIWGLFVPEE102 pKa = 4.04QVPSEE107 pKa = 4.05RR108 pKa = 11.84SEE110 pKa = 3.67PRR112 pKa = 11.84YY113 pKa = 8.65TFDD116 pKa = 3.93KK117 pKa = 10.87LSGVAPRR124 pKa = 11.84GGVKK128 pKa = 9.95KK129 pKa = 10.54QEE131 pKa = 4.03FMLHH135 pKa = 5.62CTNSSHH141 pKa = 6.64HH142 pKa = 6.91KK143 pKa = 9.8SRR145 pKa = 11.84PVLPLLLFRR154 pKa = 11.84PEE156 pKa = 4.0RR157 pKa = 11.84NSVRR161 pKa = 11.84EE162 pKa = 4.18TCCLHH167 pKa = 6.23PRR169 pKa = 11.84RR170 pKa = 11.84GRR172 pKa = 11.84KK173 pKa = 9.3AISRR177 pKa = 3.94
MM1 pKa = 7.22GRR3 pKa = 11.84SPQVQGGLKK12 pKa = 9.26GQWGSLSLLPLCCSTVACTCVSAATFFWSNFSRR45 pKa = 11.84TCTSSPAGVKK55 pKa = 10.23RR56 pKa = 11.84ILVQYY61 pKa = 10.1NISLHH66 pKa = 6.33LFVLRR71 pKa = 11.84PSAYY75 pKa = 9.57FSAGFSFVEE84 pKa = 4.23ARR86 pKa = 11.84SKK88 pKa = 10.35FARR91 pKa = 11.84ARR93 pKa = 11.84AIWGLFVPEE102 pKa = 4.04QVPSEE107 pKa = 4.05RR108 pKa = 11.84SEE110 pKa = 3.67PRR112 pKa = 11.84YY113 pKa = 8.65TFDD116 pKa = 3.93KK117 pKa = 10.87LSGVAPRR124 pKa = 11.84GGVKK128 pKa = 9.95KK129 pKa = 10.54QEE131 pKa = 4.03FMLHH135 pKa = 5.62CTNSSHH141 pKa = 6.64HH142 pKa = 6.91KK143 pKa = 9.8SRR145 pKa = 11.84PVLPLLLFRR154 pKa = 11.84PEE156 pKa = 4.0RR157 pKa = 11.84NSVRR161 pKa = 11.84EE162 pKa = 4.18TCCLHH167 pKa = 6.23PRR169 pKa = 11.84RR170 pKa = 11.84GRR172 pKa = 11.84KK173 pKa = 9.3AISRR177 pKa = 3.94
Molecular weight: 19.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1822202 |
26 |
5781 |
372.7 |
41.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.404 ± 0.037 | 0.933 ± 0.023 |
6.117 ± 0.043 | 6.402 ± 0.033 |
4.657 ± 0.027 | 7.87 ± 0.043 |
1.79 ± 0.019 | 5.66 ± 0.025 |
4.387 ± 0.048 | 10.003 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.159 ± 0.018 | 4.399 ± 0.029 |
4.579 ± 0.028 | 3.535 ± 0.021 |
5.482 ± 0.033 | 6.187 ± 0.031 |
6.036 ± 0.05 | 6.546 ± 0.024 |
1.284 ± 0.016 | 3.568 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
