
Duck faeces associated circular DNA virus 1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.17
Get precalculated fractions of proteins
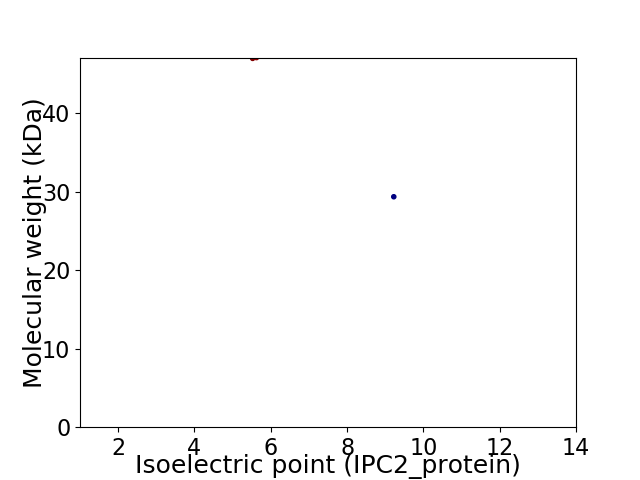
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
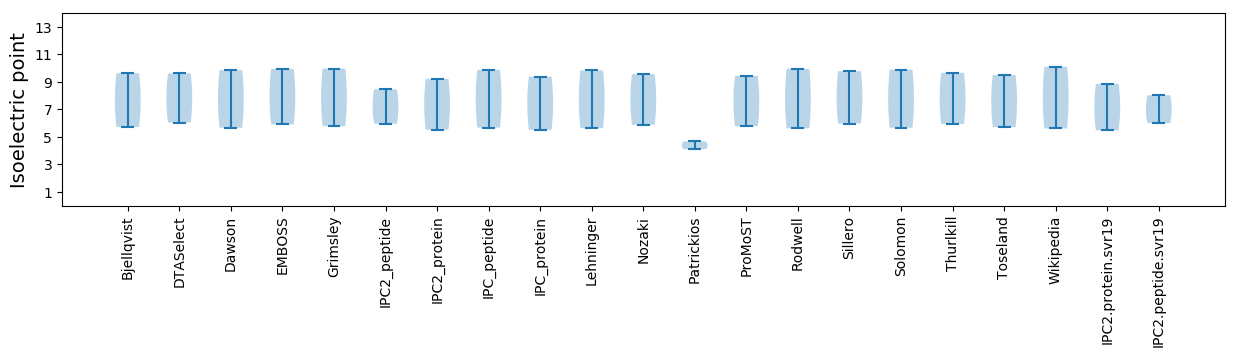
Protein with the lowest isoelectric point:
>tr|A0A161J5M7|A0A161J5M7_9VIRU Putative capsid protein OS=Duck faeces associated circular DNA virus 1 OX=1843768 PE=4 SV=1
MM1 pKa = 8.02PSFQINSKK9 pKa = 10.33SLFLTFPQCQTSLLRR24 pKa = 11.84FQEE27 pKa = 4.47LINGFFGKK35 pKa = 10.02NLEE38 pKa = 4.48KK39 pKa = 10.87GVICQEE45 pKa = 3.89NHH47 pKa = 7.03KK48 pKa = 10.65DD49 pKa = 3.71SQDD52 pKa = 3.48LHH54 pKa = 7.72LHH56 pKa = 6.09AAICLKK62 pKa = 9.41EE63 pKa = 4.23TLRR66 pKa = 11.84SRR68 pKa = 11.84DD69 pKa = 3.49PRR71 pKa = 11.84VFDD74 pKa = 4.51CLVEE78 pKa = 4.1PAQHH82 pKa = 6.59GDD84 pKa = 2.58IEE86 pKa = 4.6TRR88 pKa = 11.84FKK90 pKa = 11.4GGVLKK95 pKa = 10.89AFQYY99 pKa = 10.89VMKK102 pKa = 10.35DD103 pKa = 3.28GNYY106 pKa = 10.3LPLGQGFDD114 pKa = 3.97LNLFMEE120 pKa = 4.4QATKK124 pKa = 10.54KK125 pKa = 10.12KK126 pKa = 8.81CTKK129 pKa = 9.73TSLILKK135 pKa = 8.96DD136 pKa = 3.43IQAGATLDD144 pKa = 3.77SLVPEE149 pKa = 4.13HH150 pKa = 7.52AEE152 pKa = 3.77YY153 pKa = 11.17LLQNLQKK160 pKa = 10.76VKK162 pKa = 10.75AYY164 pKa = 10.97LDD166 pKa = 3.73FLSLRR171 pKa = 11.84EE172 pKa = 3.91KK173 pKa = 10.58RR174 pKa = 11.84LAFAEE179 pKa = 4.43GQTQKK184 pKa = 10.05VHH186 pKa = 5.41VMPAEE191 pKa = 5.4GYY193 pKa = 9.07CLSWNQEE200 pKa = 3.36IASWLNLNLRR210 pKa = 11.84QKK212 pKa = 10.36RR213 pKa = 11.84SHH215 pKa = 6.06RR216 pKa = 11.84QKK218 pKa = 11.2QMWICAPPGMGKK230 pKa = 8.33TSMIMMLEE238 pKa = 3.96KK239 pKa = 10.39AYY241 pKa = 10.45DD242 pKa = 3.6LQVYY246 pKa = 7.16YY247 pKa = 9.92WPKK250 pKa = 10.78EE251 pKa = 4.09EE252 pKa = 4.32VWWDD256 pKa = 3.44AYY258 pKa = 11.62SDD260 pKa = 3.81GAYY263 pKa = 10.64DD264 pKa = 6.07LIVLDD269 pKa = 4.8EE270 pKa = 4.25FHH272 pKa = 6.47SQKK275 pKa = 9.92TITQLNPILSGDD287 pKa = 3.7PTPLSRR293 pKa = 11.84RR294 pKa = 11.84CSAPLTKK301 pKa = 9.44RR302 pKa = 11.84DD303 pKa = 3.95NLPVIIMSNYY313 pKa = 9.7LPEE316 pKa = 4.14EE317 pKa = 4.45CFSKK321 pKa = 10.87VAANAPSKK329 pKa = 10.28LAPLIDD335 pKa = 3.69RR336 pKa = 11.84LTIVNVQGPIRR347 pKa = 11.84MAPVLEE353 pKa = 5.25DD354 pKa = 4.05DD355 pKa = 4.14PQEE358 pKa = 4.05VLAALTDD365 pKa = 3.58EE366 pKa = 4.33TSYY369 pKa = 11.58VQLTPSPGTPNLRR382 pKa = 11.84QFGDD386 pKa = 4.24DD387 pKa = 3.64EE388 pKa = 4.73EE389 pKa = 5.42DD390 pKa = 3.06WFAEE394 pKa = 4.16EE395 pKa = 4.06EE396 pKa = 4.05FRR398 pKa = 11.84RR399 pKa = 11.84NNDD402 pKa = 2.7FNLYY406 pKa = 10.1NKK408 pKa = 9.88VSII411 pKa = 4.48
MM1 pKa = 8.02PSFQINSKK9 pKa = 10.33SLFLTFPQCQTSLLRR24 pKa = 11.84FQEE27 pKa = 4.47LINGFFGKK35 pKa = 10.02NLEE38 pKa = 4.48KK39 pKa = 10.87GVICQEE45 pKa = 3.89NHH47 pKa = 7.03KK48 pKa = 10.65DD49 pKa = 3.71SQDD52 pKa = 3.48LHH54 pKa = 7.72LHH56 pKa = 6.09AAICLKK62 pKa = 9.41EE63 pKa = 4.23TLRR66 pKa = 11.84SRR68 pKa = 11.84DD69 pKa = 3.49PRR71 pKa = 11.84VFDD74 pKa = 4.51CLVEE78 pKa = 4.1PAQHH82 pKa = 6.59GDD84 pKa = 2.58IEE86 pKa = 4.6TRR88 pKa = 11.84FKK90 pKa = 11.4GGVLKK95 pKa = 10.89AFQYY99 pKa = 10.89VMKK102 pKa = 10.35DD103 pKa = 3.28GNYY106 pKa = 10.3LPLGQGFDD114 pKa = 3.97LNLFMEE120 pKa = 4.4QATKK124 pKa = 10.54KK125 pKa = 10.12KK126 pKa = 8.81CTKK129 pKa = 9.73TSLILKK135 pKa = 8.96DD136 pKa = 3.43IQAGATLDD144 pKa = 3.77SLVPEE149 pKa = 4.13HH150 pKa = 7.52AEE152 pKa = 3.77YY153 pKa = 11.17LLQNLQKK160 pKa = 10.76VKK162 pKa = 10.75AYY164 pKa = 10.97LDD166 pKa = 3.73FLSLRR171 pKa = 11.84EE172 pKa = 3.91KK173 pKa = 10.58RR174 pKa = 11.84LAFAEE179 pKa = 4.43GQTQKK184 pKa = 10.05VHH186 pKa = 5.41VMPAEE191 pKa = 5.4GYY193 pKa = 9.07CLSWNQEE200 pKa = 3.36IASWLNLNLRR210 pKa = 11.84QKK212 pKa = 10.36RR213 pKa = 11.84SHH215 pKa = 6.06RR216 pKa = 11.84QKK218 pKa = 11.2QMWICAPPGMGKK230 pKa = 8.33TSMIMMLEE238 pKa = 3.96KK239 pKa = 10.39AYY241 pKa = 10.45DD242 pKa = 3.6LQVYY246 pKa = 7.16YY247 pKa = 9.92WPKK250 pKa = 10.78EE251 pKa = 4.09EE252 pKa = 4.32VWWDD256 pKa = 3.44AYY258 pKa = 11.62SDD260 pKa = 3.81GAYY263 pKa = 10.64DD264 pKa = 6.07LIVLDD269 pKa = 4.8EE270 pKa = 4.25FHH272 pKa = 6.47SQKK275 pKa = 9.92TITQLNPILSGDD287 pKa = 3.7PTPLSRR293 pKa = 11.84RR294 pKa = 11.84CSAPLTKK301 pKa = 9.44RR302 pKa = 11.84DD303 pKa = 3.95NLPVIIMSNYY313 pKa = 9.7LPEE316 pKa = 4.14EE317 pKa = 4.45CFSKK321 pKa = 10.87VAANAPSKK329 pKa = 10.28LAPLIDD335 pKa = 3.69RR336 pKa = 11.84LTIVNVQGPIRR347 pKa = 11.84MAPVLEE353 pKa = 5.25DD354 pKa = 4.05DD355 pKa = 4.14PQEE358 pKa = 4.05VLAALTDD365 pKa = 3.58EE366 pKa = 4.33TSYY369 pKa = 11.58VQLTPSPGTPNLRR382 pKa = 11.84QFGDD386 pKa = 4.24DD387 pKa = 3.64EE388 pKa = 4.73EE389 pKa = 5.42DD390 pKa = 3.06WFAEE394 pKa = 4.16EE395 pKa = 4.06EE396 pKa = 4.05FRR398 pKa = 11.84RR399 pKa = 11.84NNDD402 pKa = 2.7FNLYY406 pKa = 10.1NKK408 pKa = 9.88VSII411 pKa = 4.48
Molecular weight: 47.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A161J5M7|A0A161J5M7_9VIRU Putative capsid protein OS=Duck faeces associated circular DNA virus 1 OX=1843768 PE=4 SV=1
MM1 pKa = 7.36LGYY4 pKa = 10.6RR5 pKa = 11.84LRR7 pKa = 11.84EE8 pKa = 4.03KK9 pKa = 10.06QAKK12 pKa = 6.55EE13 pKa = 3.61ARR15 pKa = 11.84GKK17 pKa = 9.82GVRR20 pKa = 11.84RR21 pKa = 11.84TGALVMRR28 pKa = 11.84QRR30 pKa = 11.84SRR32 pKa = 11.84LAASSVARR40 pKa = 11.84MPAYY44 pKa = 9.54IRR46 pKa = 11.84PTTNIYY52 pKa = 9.66QRR54 pKa = 11.84SGDD57 pKa = 3.94KK58 pKa = 10.7KK59 pKa = 11.43GMDD62 pKa = 3.5TDD64 pKa = 3.44ILGVSTIASATASSAGVGVLNLIQTGSGSWNRR96 pKa = 11.84VGRR99 pKa = 11.84KK100 pKa = 8.89IFPKK104 pKa = 9.9SVRR107 pKa = 11.84LRR109 pKa = 11.84YY110 pKa = 10.25GIDD113 pKa = 3.19YY114 pKa = 8.41TYY116 pKa = 11.49SNDD119 pKa = 3.55GDD121 pKa = 3.64QTYY124 pKa = 9.47PLSVRR129 pKa = 11.84VLLVWDD135 pKa = 4.85ANPSGGAKK143 pKa = 10.13PLFTEE148 pKa = 4.19MFKK151 pKa = 10.51DD152 pKa = 3.48TDD154 pKa = 3.43QTGAEE159 pKa = 4.25TTSWRR164 pKa = 11.84SGPAYY169 pKa = 10.31DD170 pKa = 3.67AMGRR174 pKa = 11.84FTILRR179 pKa = 11.84DD180 pKa = 3.48KK181 pKa = 11.13VHH183 pKa = 6.17TFVPEE188 pKa = 3.99VFPLPVTNGLARR200 pKa = 11.84SRR202 pKa = 11.84IDD204 pKa = 3.05CDD206 pKa = 3.19EE207 pKa = 4.2FVPLQQAMEE216 pKa = 4.33TVYY219 pKa = 10.69SGQNNPMTIADD230 pKa = 4.12IYY232 pKa = 11.21SGALYY237 pKa = 9.94IVSICDD243 pKa = 3.24SSPPIGYY250 pKa = 9.75GIQFSPYY257 pKa = 7.37SWARR261 pKa = 11.84LRR263 pKa = 11.84YY264 pKa = 7.82TDD266 pKa = 3.41
MM1 pKa = 7.36LGYY4 pKa = 10.6RR5 pKa = 11.84LRR7 pKa = 11.84EE8 pKa = 4.03KK9 pKa = 10.06QAKK12 pKa = 6.55EE13 pKa = 3.61ARR15 pKa = 11.84GKK17 pKa = 9.82GVRR20 pKa = 11.84RR21 pKa = 11.84TGALVMRR28 pKa = 11.84QRR30 pKa = 11.84SRR32 pKa = 11.84LAASSVARR40 pKa = 11.84MPAYY44 pKa = 9.54IRR46 pKa = 11.84PTTNIYY52 pKa = 9.66QRR54 pKa = 11.84SGDD57 pKa = 3.94KK58 pKa = 10.7KK59 pKa = 11.43GMDD62 pKa = 3.5TDD64 pKa = 3.44ILGVSTIASATASSAGVGVLNLIQTGSGSWNRR96 pKa = 11.84VGRR99 pKa = 11.84KK100 pKa = 8.89IFPKK104 pKa = 9.9SVRR107 pKa = 11.84LRR109 pKa = 11.84YY110 pKa = 10.25GIDD113 pKa = 3.19YY114 pKa = 8.41TYY116 pKa = 11.49SNDD119 pKa = 3.55GDD121 pKa = 3.64QTYY124 pKa = 9.47PLSVRR129 pKa = 11.84VLLVWDD135 pKa = 4.85ANPSGGAKK143 pKa = 10.13PLFTEE148 pKa = 4.19MFKK151 pKa = 10.51DD152 pKa = 3.48TDD154 pKa = 3.43QTGAEE159 pKa = 4.25TTSWRR164 pKa = 11.84SGPAYY169 pKa = 10.31DD170 pKa = 3.67AMGRR174 pKa = 11.84FTILRR179 pKa = 11.84DD180 pKa = 3.48KK181 pKa = 11.13VHH183 pKa = 6.17TFVPEE188 pKa = 3.99VFPLPVTNGLARR200 pKa = 11.84SRR202 pKa = 11.84IDD204 pKa = 3.05CDD206 pKa = 3.19EE207 pKa = 4.2FVPLQQAMEE216 pKa = 4.33TVYY219 pKa = 10.69SGQNNPMTIADD230 pKa = 4.12IYY232 pKa = 11.21SGALYY237 pKa = 9.94IVSICDD243 pKa = 3.24SSPPIGYY250 pKa = 9.75GIQFSPYY257 pKa = 7.37SWARR261 pKa = 11.84LRR263 pKa = 11.84YY264 pKa = 7.82TDD266 pKa = 3.41
Molecular weight: 29.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
677 |
266 |
411 |
338.5 |
38.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.942 ± 0.536 | 1.625 ± 0.492 |
6.056 ± 0.023 | 5.022 ± 1.346 |
3.988 ± 0.552 | 6.352 ± 1.504 |
1.329 ± 0.537 | 5.022 ± 0.347 |
5.465 ± 0.961 | 10.044 ± 1.846 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.806 ± 0.113 | 4.136 ± 0.635 |
5.761 ± 0.068 | 5.318 ± 0.877 |
6.056 ± 1.459 | 7.09 ± 1.088 |
5.761 ± 0.99 | 5.613 ± 0.65 |
1.625 ± 0.068 | 3.988 ± 0.718 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
