
Circovirus-like genome DCCV-13
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.56
Get precalculated fractions of proteins
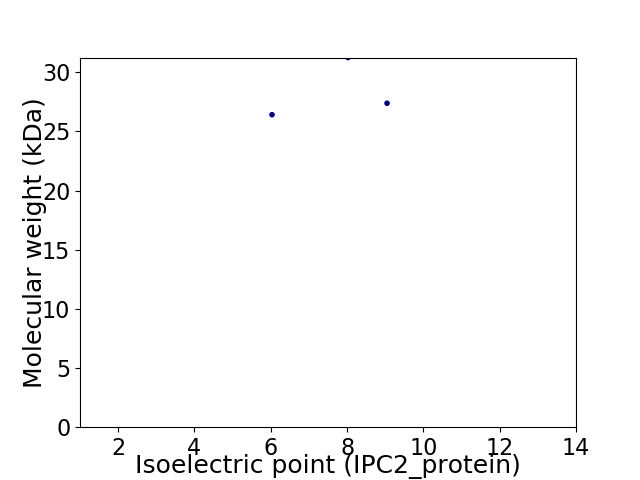
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
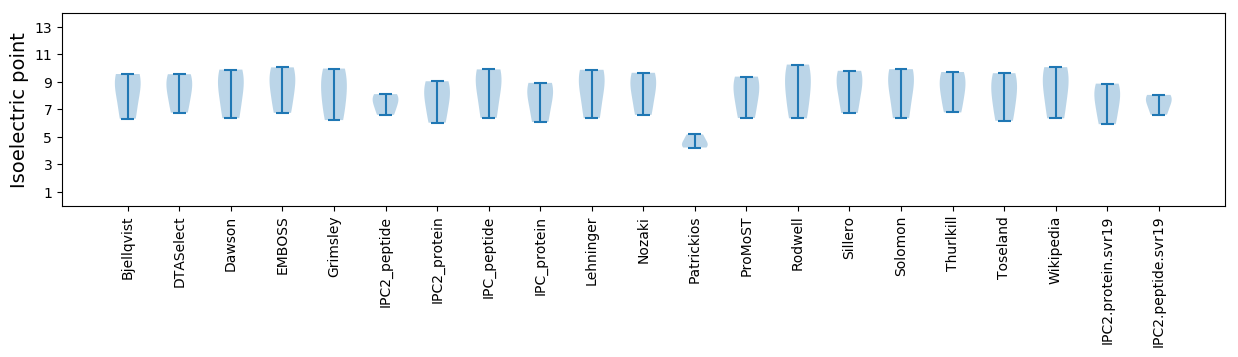
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A190WHD3|A0A190WHD3_9CIRC Replication-associated protein OS=Circovirus-like genome DCCV-13 OX=1788441 PE=4 SV=1
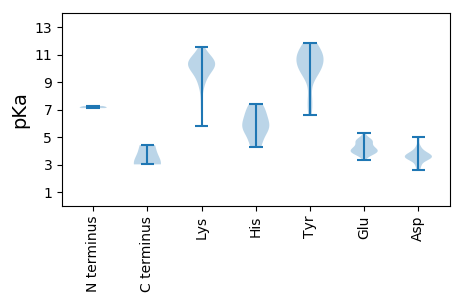
MM1 pKa = 7.26EE2 pKa = 4.86WEE4 pKa = 4.93LPNEE8 pKa = 4.45LDD10 pKa = 3.8TGCLPSPTKK19 pKa = 10.33IGPQRR24 pKa = 11.84SCSHH28 pKa = 6.68PACRR32 pKa = 11.84TYY34 pKa = 10.52EE35 pKa = 3.99VKK37 pKa = 10.29PRR39 pKa = 11.84SVQQGSNTGSCLRR52 pKa = 11.84SIPSRR57 pKa = 11.84SDD59 pKa = 3.34SLPLSPALSDD69 pKa = 2.99RR70 pKa = 11.84HH71 pKa = 5.64TANQPDD77 pKa = 4.34QKK79 pKa = 11.01PPMTMFGRR87 pKa = 11.84KK88 pKa = 5.78TPRR91 pKa = 11.84WTEE94 pKa = 3.57QDD96 pKa = 3.87LKK98 pKa = 10.49WVNDD102 pKa = 3.55LTEE105 pKa = 4.55EE106 pKa = 4.09MSKK109 pKa = 8.59PTGSEE114 pKa = 3.72YY115 pKa = 11.16GNPLLQGTFCPSKK128 pKa = 10.41QVFVFNITAHH138 pKa = 5.33SVQYY142 pKa = 11.6ALTTLNQLLLNVEE155 pKa = 4.64SLSFMDD161 pKa = 5.0LQVLEE166 pKa = 4.26NRR168 pKa = 11.84EE169 pKa = 4.15EE170 pKa = 4.19PGKK173 pKa = 10.91KK174 pKa = 9.45PVGLLILKK182 pKa = 9.73IRR184 pKa = 11.84EE185 pKa = 4.28ANFGMDD191 pKa = 3.3TQIRR195 pKa = 11.84RR196 pKa = 11.84MLYY199 pKa = 10.04LMNFEE204 pKa = 4.79EE205 pKa = 4.71EE206 pKa = 4.71SIFPIYY212 pKa = 10.26CDD214 pKa = 3.57GLIATQYY221 pKa = 9.56SLKK224 pKa = 10.59SRR226 pKa = 11.84EE227 pKa = 4.18LQRR230 pKa = 11.84VWW232 pKa = 3.02
MM1 pKa = 7.26EE2 pKa = 4.86WEE4 pKa = 4.93LPNEE8 pKa = 4.45LDD10 pKa = 3.8TGCLPSPTKK19 pKa = 10.33IGPQRR24 pKa = 11.84SCSHH28 pKa = 6.68PACRR32 pKa = 11.84TYY34 pKa = 10.52EE35 pKa = 3.99VKK37 pKa = 10.29PRR39 pKa = 11.84SVQQGSNTGSCLRR52 pKa = 11.84SIPSRR57 pKa = 11.84SDD59 pKa = 3.34SLPLSPALSDD69 pKa = 2.99RR70 pKa = 11.84HH71 pKa = 5.64TANQPDD77 pKa = 4.34QKK79 pKa = 11.01PPMTMFGRR87 pKa = 11.84KK88 pKa = 5.78TPRR91 pKa = 11.84WTEE94 pKa = 3.57QDD96 pKa = 3.87LKK98 pKa = 10.49WVNDD102 pKa = 3.55LTEE105 pKa = 4.55EE106 pKa = 4.09MSKK109 pKa = 8.59PTGSEE114 pKa = 3.72YY115 pKa = 11.16GNPLLQGTFCPSKK128 pKa = 10.41QVFVFNITAHH138 pKa = 5.33SVQYY142 pKa = 11.6ALTTLNQLLLNVEE155 pKa = 4.64SLSFMDD161 pKa = 5.0LQVLEE166 pKa = 4.26NRR168 pKa = 11.84EE169 pKa = 4.15EE170 pKa = 4.19PGKK173 pKa = 10.91KK174 pKa = 9.45PVGLLILKK182 pKa = 9.73IRR184 pKa = 11.84EE185 pKa = 4.28ANFGMDD191 pKa = 3.3TQIRR195 pKa = 11.84RR196 pKa = 11.84MLYY199 pKa = 10.04LMNFEE204 pKa = 4.79EE205 pKa = 4.71EE206 pKa = 4.71SIFPIYY212 pKa = 10.26CDD214 pKa = 3.57GLIATQYY221 pKa = 9.56SLKK224 pKa = 10.59SRR226 pKa = 11.84EE227 pKa = 4.18LQRR230 pKa = 11.84VWW232 pKa = 3.02
Molecular weight: 26.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A190WHE0|A0A190WHE0_9CIRC Uncharacterized protein OS=Circovirus-like genome DCCV-13 OX=1788441 PE=4 SV=1
MM1 pKa = 7.17AVKK4 pKa = 10.37RR5 pKa = 11.84KK6 pKa = 8.59MPQSQGKK13 pKa = 7.72YY14 pKa = 6.58VKK16 pKa = 9.77KK17 pKa = 10.56RR18 pKa = 11.84KK19 pKa = 6.92STKK22 pKa = 9.31VNKK25 pKa = 10.11LQVKK29 pKa = 10.09KK30 pKa = 10.09MILGLSEE37 pKa = 4.01TKK39 pKa = 9.89HH40 pKa = 4.35YY41 pKa = 10.69HH42 pKa = 4.26STYY45 pKa = 9.36VAEE48 pKa = 4.64IKK50 pKa = 9.81TGFQYY55 pKa = 10.99NINPLYY61 pKa = 9.68WIPIGSNEE69 pKa = 3.95TSRR72 pKa = 11.84LGDD75 pKa = 3.61KK76 pKa = 10.6IFLEE80 pKa = 4.84SIDD83 pKa = 4.22MNCKK87 pKa = 9.03IDD89 pKa = 3.7RR90 pKa = 11.84SNVDD94 pKa = 2.74VGGLGRR100 pKa = 11.84FKK102 pKa = 10.98NSSIPFNLKK111 pKa = 10.47LIRR114 pKa = 11.84CTSKK118 pKa = 10.85AKK120 pKa = 10.52QGTTGDD126 pKa = 3.34AGFTLLGLPDD136 pKa = 3.57IRR138 pKa = 11.84LDD140 pKa = 3.87ALGNFANPHH149 pKa = 5.73NNLQQFQVVWDD160 pKa = 3.72YY161 pKa = 11.68RR162 pKa = 11.84GTLEE166 pKa = 5.3QMPVLNAGNDD176 pKa = 3.5QYY178 pKa = 11.82NSVMIKK184 pKa = 10.14KK185 pKa = 9.92RR186 pKa = 11.84IPINRR191 pKa = 11.84QVLYY195 pKa = 10.66DD196 pKa = 3.58GVEE199 pKa = 4.0ITGSSGFLKK208 pKa = 10.36DD209 pKa = 3.47YY210 pKa = 10.86QYY212 pKa = 11.81YY213 pKa = 10.07FYY215 pKa = 10.86FAVDD219 pKa = 3.56SNTTAGLSAVFVHH232 pKa = 7.18SDD234 pKa = 2.6ILVNFKK240 pKa = 11.31DD241 pKa = 3.57MM242 pKa = 4.41
MM1 pKa = 7.17AVKK4 pKa = 10.37RR5 pKa = 11.84KK6 pKa = 8.59MPQSQGKK13 pKa = 7.72YY14 pKa = 6.58VKK16 pKa = 9.77KK17 pKa = 10.56RR18 pKa = 11.84KK19 pKa = 6.92STKK22 pKa = 9.31VNKK25 pKa = 10.11LQVKK29 pKa = 10.09KK30 pKa = 10.09MILGLSEE37 pKa = 4.01TKK39 pKa = 9.89HH40 pKa = 4.35YY41 pKa = 10.69HH42 pKa = 4.26STYY45 pKa = 9.36VAEE48 pKa = 4.64IKK50 pKa = 9.81TGFQYY55 pKa = 10.99NINPLYY61 pKa = 9.68WIPIGSNEE69 pKa = 3.95TSRR72 pKa = 11.84LGDD75 pKa = 3.61KK76 pKa = 10.6IFLEE80 pKa = 4.84SIDD83 pKa = 4.22MNCKK87 pKa = 9.03IDD89 pKa = 3.7RR90 pKa = 11.84SNVDD94 pKa = 2.74VGGLGRR100 pKa = 11.84FKK102 pKa = 10.98NSSIPFNLKK111 pKa = 10.47LIRR114 pKa = 11.84CTSKK118 pKa = 10.85AKK120 pKa = 10.52QGTTGDD126 pKa = 3.34AGFTLLGLPDD136 pKa = 3.57IRR138 pKa = 11.84LDD140 pKa = 3.87ALGNFANPHH149 pKa = 5.73NNLQQFQVVWDD160 pKa = 3.72YY161 pKa = 11.68RR162 pKa = 11.84GTLEE166 pKa = 5.3QMPVLNAGNDD176 pKa = 3.5QYY178 pKa = 11.82NSVMIKK184 pKa = 10.14KK185 pKa = 9.92RR186 pKa = 11.84IPINRR191 pKa = 11.84QVLYY195 pKa = 10.66DD196 pKa = 3.58GVEE199 pKa = 4.0ITGSSGFLKK208 pKa = 10.36DD209 pKa = 3.47YY210 pKa = 10.86QYY212 pKa = 11.81YY213 pKa = 10.07FYY215 pKa = 10.86FAVDD219 pKa = 3.56SNTTAGLSAVFVHH232 pKa = 7.18SDD234 pKa = 2.6ILVNFKK240 pKa = 11.31DD241 pKa = 3.57MM242 pKa = 4.41
Molecular weight: 27.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
742 |
232 |
268 |
247.3 |
28.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.987 ± 1.105 | 1.348 ± 0.475 |
5.391 ± 0.593 | 5.526 ± 1.261 |
3.908 ± 0.434 | 6.334 ± 0.648 |
2.022 ± 0.427 | 5.93 ± 0.793 |
6.199 ± 1.193 | 8.491 ± 1.415 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.426 ± 0.582 | 4.447 ± 1.466 |
5.526 ± 1.133 | 4.447 ± 0.832 |
6.604 ± 1.294 | 7.278 ± 0.724 |
6.469 ± 0.456 | 6.065 ± 0.678 |
2.426 ± 0.934 | 4.178 ± 0.611 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
