
Fly associated circular virus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; unclassified Porprismacovirus
Average proteome isoelectric point is 7.7
Get precalculated fractions of proteins
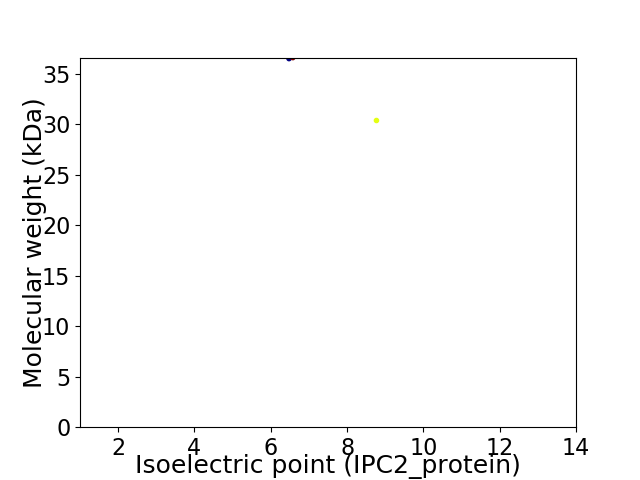
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPB3|A0A346BPB3_9VIRU Putative capsid protein OS=Fly associated circular virus 3 OX=2293283 PE=4 SV=1
MM1 pKa = 8.09AYY3 pKa = 10.04RR4 pKa = 11.84KK5 pKa = 9.62RR6 pKa = 11.84RR7 pKa = 11.84GSKK10 pKa = 9.55VLHH13 pKa = 6.49QKK15 pKa = 8.52FQWFFDD21 pKa = 3.94LQTKK25 pKa = 9.57ADD27 pKa = 3.7EE28 pKa = 4.12MQIIEE33 pKa = 4.4VSAGGYY39 pKa = 9.27GVYY42 pKa = 10.2KK43 pKa = 10.55RR44 pKa = 11.84LFPFFSAFKK53 pKa = 8.75YY54 pKa = 10.37YY55 pKa = 10.95KK56 pKa = 10.56LGGVKK61 pKa = 9.37MKK63 pKa = 9.33MIPASTLPVDD73 pKa = 3.87PTGLSYY79 pKa = 10.68EE80 pKa = 4.23AGEE83 pKa = 4.26NTVDD87 pKa = 4.85PRR89 pKa = 11.84DD90 pKa = 3.48QLTPGLTRR98 pKa = 11.84ITNGEE103 pKa = 4.0DD104 pKa = 3.58VYY106 pKa = 11.46TKK108 pKa = 10.95LDD110 pKa = 3.79GLTGDD115 pKa = 3.77QQRR118 pKa = 11.84EE119 pKa = 4.43LYY121 pKa = 9.6EE122 pKa = 4.92SMMIDD127 pKa = 3.25QRR129 pKa = 11.84WFKK132 pKa = 10.13WQLQSGLSRR141 pKa = 11.84YY142 pKa = 8.4ARR144 pKa = 11.84PMYY147 pKa = 8.75WQIGQLHH154 pKa = 6.12QDD156 pKa = 3.76YY157 pKa = 11.0LPGSVRR163 pKa = 11.84NLADD167 pKa = 3.42TNLTEE172 pKa = 4.73DD173 pKa = 4.56CDD175 pKa = 4.77SISAVYY181 pKa = 10.52DD182 pKa = 3.37GANPSGPSAGPITVAADD199 pKa = 3.17ASDD202 pKa = 3.55PRR204 pKa = 11.84GLFQVGHH211 pKa = 6.51RR212 pKa = 11.84GKK214 pKa = 10.35LGWMPTDD221 pKa = 3.49GVLLKK226 pKa = 9.81GTSGGDD232 pKa = 3.59GTVHH236 pKa = 5.98AQAVEE241 pKa = 3.92AAIPAVNVFTIVTPPMHH258 pKa = 6.15KK259 pKa = 8.5TNYY262 pKa = 7.64YY263 pKa = 8.29YY264 pKa = 10.56RR265 pKa = 11.84VFVTEE270 pKa = 4.34DD271 pKa = 3.84VYY273 pKa = 11.28FKK275 pKa = 11.2SPVVVGYY282 pKa = 10.0QNFRR286 pKa = 11.84SLDD289 pKa = 3.48RR290 pKa = 11.84FVQPQFPVAKK300 pKa = 9.97LPTVNSATTDD310 pKa = 3.39SNKK313 pKa = 9.75PFPTNDD319 pKa = 3.19GGEE322 pKa = 4.25LPGVIQQ328 pKa = 4.56
MM1 pKa = 8.09AYY3 pKa = 10.04RR4 pKa = 11.84KK5 pKa = 9.62RR6 pKa = 11.84RR7 pKa = 11.84GSKK10 pKa = 9.55VLHH13 pKa = 6.49QKK15 pKa = 8.52FQWFFDD21 pKa = 3.94LQTKK25 pKa = 9.57ADD27 pKa = 3.7EE28 pKa = 4.12MQIIEE33 pKa = 4.4VSAGGYY39 pKa = 9.27GVYY42 pKa = 10.2KK43 pKa = 10.55RR44 pKa = 11.84LFPFFSAFKK53 pKa = 8.75YY54 pKa = 10.37YY55 pKa = 10.95KK56 pKa = 10.56LGGVKK61 pKa = 9.37MKK63 pKa = 9.33MIPASTLPVDD73 pKa = 3.87PTGLSYY79 pKa = 10.68EE80 pKa = 4.23AGEE83 pKa = 4.26NTVDD87 pKa = 4.85PRR89 pKa = 11.84DD90 pKa = 3.48QLTPGLTRR98 pKa = 11.84ITNGEE103 pKa = 4.0DD104 pKa = 3.58VYY106 pKa = 11.46TKK108 pKa = 10.95LDD110 pKa = 3.79GLTGDD115 pKa = 3.77QQRR118 pKa = 11.84EE119 pKa = 4.43LYY121 pKa = 9.6EE122 pKa = 4.92SMMIDD127 pKa = 3.25QRR129 pKa = 11.84WFKK132 pKa = 10.13WQLQSGLSRR141 pKa = 11.84YY142 pKa = 8.4ARR144 pKa = 11.84PMYY147 pKa = 8.75WQIGQLHH154 pKa = 6.12QDD156 pKa = 3.76YY157 pKa = 11.0LPGSVRR163 pKa = 11.84NLADD167 pKa = 3.42TNLTEE172 pKa = 4.73DD173 pKa = 4.56CDD175 pKa = 4.77SISAVYY181 pKa = 10.52DD182 pKa = 3.37GANPSGPSAGPITVAADD199 pKa = 3.17ASDD202 pKa = 3.55PRR204 pKa = 11.84GLFQVGHH211 pKa = 6.51RR212 pKa = 11.84GKK214 pKa = 10.35LGWMPTDD221 pKa = 3.49GVLLKK226 pKa = 9.81GTSGGDD232 pKa = 3.59GTVHH236 pKa = 5.98AQAVEE241 pKa = 3.92AAIPAVNVFTIVTPPMHH258 pKa = 6.15KK259 pKa = 8.5TNYY262 pKa = 7.64YY263 pKa = 8.29YY264 pKa = 10.56RR265 pKa = 11.84VFVTEE270 pKa = 4.34DD271 pKa = 3.84VYY273 pKa = 11.28FKK275 pKa = 11.2SPVVVGYY282 pKa = 10.0QNFRR286 pKa = 11.84SLDD289 pKa = 3.48RR290 pKa = 11.84FVQPQFPVAKK300 pKa = 9.97LPTVNSATTDD310 pKa = 3.39SNKK313 pKa = 9.75PFPTNDD319 pKa = 3.19GGEE322 pKa = 4.25LPGVIQQ328 pKa = 4.56
Molecular weight: 36.48 kDa
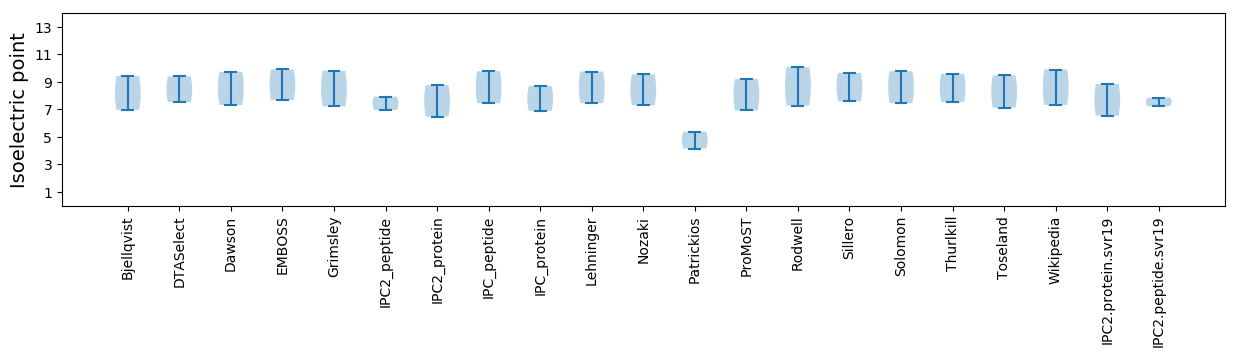
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPB3|A0A346BPB3_9VIRU Putative capsid protein OS=Fly associated circular virus 3 OX=2293283 PE=4 SV=1
MM1 pKa = 8.06DD2 pKa = 4.29AQRR5 pKa = 11.84RR6 pKa = 11.84YY7 pKa = 10.56KK8 pKa = 10.24FRR10 pKa = 11.84HH11 pKa = 4.91TLASTMTQTWMLTVPRR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 7.48TAKK32 pKa = 9.2EE33 pKa = 3.31WMGIYY38 pKa = 9.82KK39 pKa = 9.73WLRR42 pKa = 11.84DD43 pKa = 3.58NDD45 pKa = 3.68VHH47 pKa = 6.67KK48 pKa = 9.25WTVAMEE54 pKa = 4.34TGSNGYY60 pKa = 8.07DD61 pKa = 2.53HH62 pKa = 6.6WQIRR66 pKa = 11.84FQVGKK71 pKa = 7.81TFKK74 pKa = 10.19QLKK77 pKa = 9.71KK78 pKa = 9.23EE79 pKa = 4.16WGDD82 pKa = 3.38KK83 pKa = 10.52AHH85 pKa = 7.06IEE87 pKa = 4.2EE88 pKa = 5.99ASDD91 pKa = 2.85TWDD94 pKa = 3.46YY95 pKa = 11.1EE96 pKa = 4.44RR97 pKa = 11.84KK98 pKa = 10.0SGMFFSSTDD107 pKa = 3.16TPEE110 pKa = 3.59VRR112 pKa = 11.84KK113 pKa = 10.24CRR115 pKa = 11.84FGRR118 pKa = 11.84LNWRR122 pKa = 11.84QEE124 pKa = 3.92AVVQAVQATNDD135 pKa = 3.52RR136 pKa = 11.84EE137 pKa = 4.43VVVWYY142 pKa = 10.18DD143 pKa = 3.17PDD145 pKa = 4.09GNKK148 pKa = 10.05GKK150 pKa = 10.18SWLLGHH156 pKa = 7.53LYY158 pKa = 8.31EE159 pKa = 5.2TGQAWVVQAQDD170 pKa = 3.45TVKK173 pKa = 11.21GIIQDD178 pKa = 3.64VASEE182 pKa = 4.4YY183 pKa = 10.39INHH186 pKa = 5.83GWRR189 pKa = 11.84PIVVIDD195 pKa = 5.32IPRR198 pKa = 11.84TWKK201 pKa = 7.87WTSDD205 pKa = 3.27LYY207 pKa = 11.39VAIEE211 pKa = 4.27RR212 pKa = 11.84IKK214 pKa = 11.05DD215 pKa = 3.64GLIKK219 pKa = 10.46DD220 pKa = 3.78PRR222 pKa = 11.84YY223 pKa = 9.95SSKK226 pKa = 9.35TVHH229 pKa = 5.5IRR231 pKa = 11.84GVKK234 pKa = 10.04VLVTCSTMPKK244 pKa = 9.65LDD246 pKa = 3.93KK247 pKa = 10.91LSADD251 pKa = 2.92RR252 pKa = 11.84WVIIDD257 pKa = 3.73LL258 pKa = 4.19
MM1 pKa = 8.06DD2 pKa = 4.29AQRR5 pKa = 11.84RR6 pKa = 11.84YY7 pKa = 10.56KK8 pKa = 10.24FRR10 pKa = 11.84HH11 pKa = 4.91TLASTMTQTWMLTVPRR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 7.48TAKK32 pKa = 9.2EE33 pKa = 3.31WMGIYY38 pKa = 9.82KK39 pKa = 9.73WLRR42 pKa = 11.84DD43 pKa = 3.58NDD45 pKa = 3.68VHH47 pKa = 6.67KK48 pKa = 9.25WTVAMEE54 pKa = 4.34TGSNGYY60 pKa = 8.07DD61 pKa = 2.53HH62 pKa = 6.6WQIRR66 pKa = 11.84FQVGKK71 pKa = 7.81TFKK74 pKa = 10.19QLKK77 pKa = 9.71KK78 pKa = 9.23EE79 pKa = 4.16WGDD82 pKa = 3.38KK83 pKa = 10.52AHH85 pKa = 7.06IEE87 pKa = 4.2EE88 pKa = 5.99ASDD91 pKa = 2.85TWDD94 pKa = 3.46YY95 pKa = 11.1EE96 pKa = 4.44RR97 pKa = 11.84KK98 pKa = 10.0SGMFFSSTDD107 pKa = 3.16TPEE110 pKa = 3.59VRR112 pKa = 11.84KK113 pKa = 10.24CRR115 pKa = 11.84FGRR118 pKa = 11.84LNWRR122 pKa = 11.84QEE124 pKa = 3.92AVVQAVQATNDD135 pKa = 3.52RR136 pKa = 11.84EE137 pKa = 4.43VVVWYY142 pKa = 10.18DD143 pKa = 3.17PDD145 pKa = 4.09GNKK148 pKa = 10.05GKK150 pKa = 10.18SWLLGHH156 pKa = 7.53LYY158 pKa = 8.31EE159 pKa = 5.2TGQAWVVQAQDD170 pKa = 3.45TVKK173 pKa = 11.21GIIQDD178 pKa = 3.64VASEE182 pKa = 4.4YY183 pKa = 10.39INHH186 pKa = 5.83GWRR189 pKa = 11.84PIVVIDD195 pKa = 5.32IPRR198 pKa = 11.84TWKK201 pKa = 7.87WTSDD205 pKa = 3.27LYY207 pKa = 11.39VAIEE211 pKa = 4.27RR212 pKa = 11.84IKK214 pKa = 11.05DD215 pKa = 3.64GLIKK219 pKa = 10.46DD220 pKa = 3.78PRR222 pKa = 11.84YY223 pKa = 9.95SSKK226 pKa = 9.35TVHH229 pKa = 5.5IRR231 pKa = 11.84GVKK234 pKa = 10.04VLVTCSTMPKK244 pKa = 9.65LDD246 pKa = 3.93KK247 pKa = 10.91LSADD251 pKa = 2.92RR252 pKa = 11.84WVIIDD257 pKa = 3.73LL258 pKa = 4.19
Molecular weight: 30.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
586 |
258 |
328 |
293.0 |
33.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.973 ± 0.323 | 0.512 ± 0.156 |
7.167 ± 0.346 | 3.925 ± 0.429 |
3.754 ± 0.844 | 7.85 ± 1.203 |
2.048 ± 0.393 | 4.437 ± 0.814 |
6.655 ± 1.106 | 6.485 ± 0.626 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.73 ± 0.01 | 2.901 ± 0.34 |
5.119 ± 1.422 | 5.29 ± 0.378 |
5.973 ± 0.823 | 5.461 ± 0.249 |
7.338 ± 0.245 | 8.532 ± 0.226 |
3.413 ± 1.419 | 4.437 ± 0.561 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
