
Streptomyces phage Forthebois
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Kalamavirales; Tectiviridae; Deltatectivirus; Streptomyces virus Forthebois
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
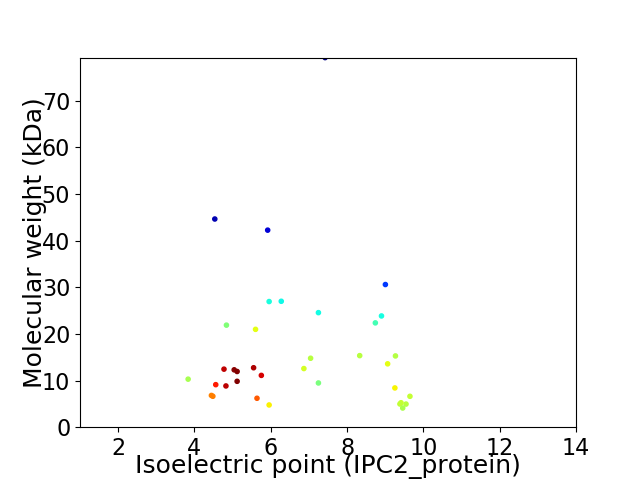
Virtual 2D-PAGE plot for 36 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A4D6E2V8|A0A4D6E2V8_9VIRU Uncharacterized protein OS=Streptomyces phage Forthebois OX=2562185 GN=1 PE=4 SV=1
MM1 pKa = 7.64EE2 pKa = 5.8FGNPIVGGEE11 pKa = 3.96DD12 pKa = 4.54LIRR15 pKa = 11.84TAIKK19 pKa = 10.81SPDD22 pKa = 3.62FNTDD26 pKa = 3.38PEE28 pKa = 4.61SGNVTGWRR36 pKa = 11.84IARR39 pKa = 11.84DD40 pKa = 3.76GSATFYY46 pKa = 11.33NLTIGSADD54 pKa = 3.58FNIDD58 pKa = 3.25EE59 pKa = 4.75NGNAVFQSVTANDD72 pKa = 3.16ITLDD76 pKa = 3.83GEE78 pKa = 4.23NLEE81 pKa = 4.41SVLAEE86 pKa = 4.13KK87 pKa = 10.82ALGTLAIVTLSGDD100 pKa = 3.41TAGYY104 pKa = 10.52NGTSLLFGRR113 pKa = 11.84IVIPNFDD120 pKa = 3.77GTRR123 pKa = 11.84QYY125 pKa = 11.5AIGGSGVHH133 pKa = 6.44FDD135 pKa = 3.56KK136 pKa = 11.23QAVTGFTRR144 pKa = 11.84LTIRR148 pKa = 11.84AYY150 pKa = 10.57LAWDD154 pKa = 4.2TPATTASTQLVEE166 pKa = 4.05YY167 pKa = 10.44QYY169 pKa = 11.62LSDD172 pKa = 4.06SASGSDD178 pKa = 3.34WIVSFRR184 pKa = 11.84HH185 pKa = 5.97LFQDD189 pKa = 3.89SSPAGTDD196 pKa = 3.2AHH198 pKa = 6.17IAWYY202 pKa = 7.81FTTNVNNTAGLRR214 pKa = 11.84CSGYY218 pKa = 10.64DD219 pKa = 3.34SGGGTPGSRR228 pKa = 11.84IYY230 pKa = 11.2AEE232 pKa = 4.18DD233 pKa = 3.62AGDD236 pKa = 3.26IVTYY240 pKa = 10.58TDD242 pKa = 3.78FNMGGGGGTPVQSYY256 pKa = 8.24TKK258 pKa = 9.38TYY260 pKa = 10.08AANEE264 pKa = 4.25SASYY268 pKa = 9.62QQDD271 pKa = 2.87GSNRR275 pKa = 11.84GIADD279 pKa = 4.53CYY281 pKa = 8.67QGRR284 pKa = 11.84YY285 pKa = 8.86SATNGNQYY293 pKa = 11.65SMIGFDD299 pKa = 3.79DD300 pKa = 3.96AQIRR304 pKa = 11.84SDD306 pKa = 3.83LSGATITKK314 pKa = 10.09VEE316 pKa = 4.29LYY318 pKa = 10.8LNNNHH323 pKa = 6.41FYY325 pKa = 11.25SNSGGNAVIGTHH337 pKa = 5.49NQTTLSGSHH346 pKa = 6.46SSSQINDD353 pKa = 3.29NLQQTHH359 pKa = 6.59FDD361 pKa = 3.43LGQAKK366 pKa = 9.85WITIPNSIGNALRR379 pKa = 11.84DD380 pKa = 3.75NTAKK384 pKa = 10.89GIALGPGPTTSQSYY398 pKa = 9.76YY399 pKa = 11.02GYY401 pKa = 10.39FAGNGQSGEE410 pKa = 3.96PQLRR414 pKa = 11.84ITYY417 pKa = 8.08TKK419 pKa = 10.94
MM1 pKa = 7.64EE2 pKa = 5.8FGNPIVGGEE11 pKa = 3.96DD12 pKa = 4.54LIRR15 pKa = 11.84TAIKK19 pKa = 10.81SPDD22 pKa = 3.62FNTDD26 pKa = 3.38PEE28 pKa = 4.61SGNVTGWRR36 pKa = 11.84IARR39 pKa = 11.84DD40 pKa = 3.76GSATFYY46 pKa = 11.33NLTIGSADD54 pKa = 3.58FNIDD58 pKa = 3.25EE59 pKa = 4.75NGNAVFQSVTANDD72 pKa = 3.16ITLDD76 pKa = 3.83GEE78 pKa = 4.23NLEE81 pKa = 4.41SVLAEE86 pKa = 4.13KK87 pKa = 10.82ALGTLAIVTLSGDD100 pKa = 3.41TAGYY104 pKa = 10.52NGTSLLFGRR113 pKa = 11.84IVIPNFDD120 pKa = 3.77GTRR123 pKa = 11.84QYY125 pKa = 11.5AIGGSGVHH133 pKa = 6.44FDD135 pKa = 3.56KK136 pKa = 11.23QAVTGFTRR144 pKa = 11.84LTIRR148 pKa = 11.84AYY150 pKa = 10.57LAWDD154 pKa = 4.2TPATTASTQLVEE166 pKa = 4.05YY167 pKa = 10.44QYY169 pKa = 11.62LSDD172 pKa = 4.06SASGSDD178 pKa = 3.34WIVSFRR184 pKa = 11.84HH185 pKa = 5.97LFQDD189 pKa = 3.89SSPAGTDD196 pKa = 3.2AHH198 pKa = 6.17IAWYY202 pKa = 7.81FTTNVNNTAGLRR214 pKa = 11.84CSGYY218 pKa = 10.64DD219 pKa = 3.34SGGGTPGSRR228 pKa = 11.84IYY230 pKa = 11.2AEE232 pKa = 4.18DD233 pKa = 3.62AGDD236 pKa = 3.26IVTYY240 pKa = 10.58TDD242 pKa = 3.78FNMGGGGGTPVQSYY256 pKa = 8.24TKK258 pKa = 9.38TYY260 pKa = 10.08AANEE264 pKa = 4.25SASYY268 pKa = 9.62QQDD271 pKa = 2.87GSNRR275 pKa = 11.84GIADD279 pKa = 4.53CYY281 pKa = 8.67QGRR284 pKa = 11.84YY285 pKa = 8.86SATNGNQYY293 pKa = 11.65SMIGFDD299 pKa = 3.79DD300 pKa = 3.96AQIRR304 pKa = 11.84SDD306 pKa = 3.83LSGATITKK314 pKa = 10.09VEE316 pKa = 4.29LYY318 pKa = 10.8LNNNHH323 pKa = 6.41FYY325 pKa = 11.25SNSGGNAVIGTHH337 pKa = 5.49NQTTLSGSHH346 pKa = 6.46SSSQINDD353 pKa = 3.29NLQQTHH359 pKa = 6.59FDD361 pKa = 3.43LGQAKK366 pKa = 9.85WITIPNSIGNALRR379 pKa = 11.84DD380 pKa = 3.75NTAKK384 pKa = 10.89GIALGPGPTTSQSYY398 pKa = 9.76YY399 pKa = 11.02GYY401 pKa = 10.39FAGNGQSGEE410 pKa = 3.96PQLRR414 pKa = 11.84ITYY417 pKa = 8.08TKK419 pKa = 10.94
Molecular weight: 44.64 kDa
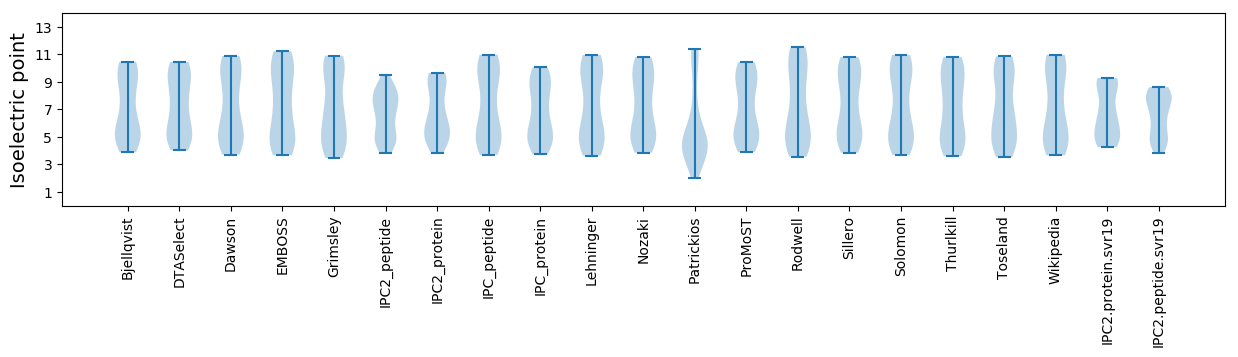
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6E2V9|A0A4D6E2V9_9VIRU Uncharacterized protein OS=Streptomyces phage Forthebois OX=2562185 GN=9 PE=4 SV=1
MM1 pKa = 7.58AAQRR5 pKa = 11.84SMKK8 pKa = 10.16DD9 pKa = 2.82KK10 pKa = 10.85MMAAMLKK17 pKa = 10.43DD18 pKa = 3.26KK19 pKa = 11.14GIARR23 pKa = 11.84TTGQCPRR30 pKa = 11.84CHH32 pKa = 6.45KK33 pKa = 10.12FAYY36 pKa = 10.04SIKK39 pKa = 9.62NQNSLILHH47 pKa = 6.65LNGCQGRR54 pKa = 11.84RR55 pKa = 11.84AKK57 pKa = 10.95VNN59 pKa = 3.23
MM1 pKa = 7.58AAQRR5 pKa = 11.84SMKK8 pKa = 10.16DD9 pKa = 2.82KK10 pKa = 10.85MMAAMLKK17 pKa = 10.43DD18 pKa = 3.26KK19 pKa = 11.14GIARR23 pKa = 11.84TTGQCPRR30 pKa = 11.84CHH32 pKa = 6.45KK33 pKa = 10.12FAYY36 pKa = 10.04SIKK39 pKa = 9.62NQNSLILHH47 pKa = 6.65LNGCQGRR54 pKa = 11.84RR55 pKa = 11.84AKK57 pKa = 10.95VNN59 pKa = 3.23
Molecular weight: 6.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5544 |
38 |
700 |
154.0 |
16.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.011 ± 0.622 | 0.685 ± 0.18 |
5.592 ± 0.281 | 4.96 ± 0.475 |
3.481 ± 0.228 | 9.578 ± 0.647 |
1.641 ± 0.189 | 5.195 ± 0.367 |
5.213 ± 0.47 | 6.836 ± 0.528 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.814 ± 0.36 | 4.816 ± 0.376 |
5.628 ± 0.558 | 3.535 ± 0.384 |
5.014 ± 0.544 | 6.025 ± 0.388 |
7.143 ± 0.545 | 6.854 ± 0.59 |
1.858 ± 0.206 | 3.12 ± 0.312 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
