
Ruegeria pomeroyi (strain ATCC 700808 / DSM 15171 / DSS-3) (Silicibacter pomeroyi)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Ruegeria; Ruegeria pomeroyi
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
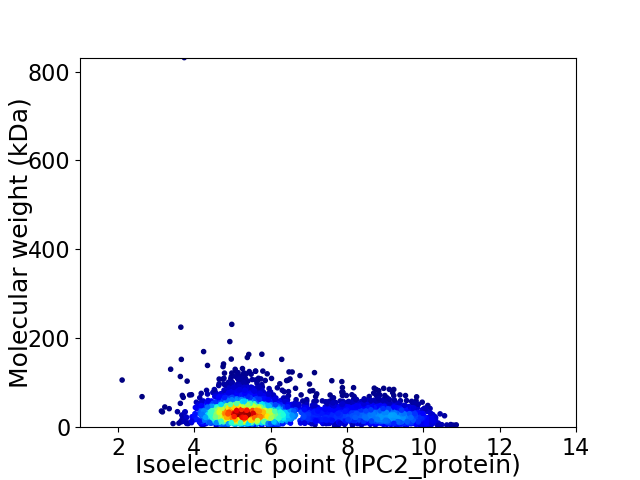
Virtual 2D-PAGE plot for 4269 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q5LNP8|Q5LNP8_RUEPO UPF0260 protein SPO3155 OS=Ruegeria pomeroyi (strain ATCC 700808 / DSM 15171 / DSS-3) OX=246200 GN=SPO3155 PE=3 SV=1
MM1 pKa = 7.43RR2 pKa = 11.84AAVRR6 pKa = 11.84LQLLPHH12 pKa = 6.62MDD14 pKa = 3.72SIYY17 pKa = 10.54FFCNLTACYY26 pKa = 10.44YY27 pKa = 11.06LLLNRR32 pKa = 11.84GTLVLGTILFAALGVAAIATVISSDD57 pKa = 3.72DD58 pKa = 3.76DD59 pKa = 3.78SHH61 pKa = 8.87EE62 pKa = 4.54EE63 pKa = 3.88IADD66 pKa = 3.66GDD68 pKa = 4.04VPPPIEE74 pKa = 4.83EE75 pKa = 4.08EE76 pKa = 4.32DD77 pKa = 3.83GTPSLIQEE85 pKa = 4.29NGLTVVTGNEE95 pKa = 4.0EE96 pKa = 3.73PVQIDD101 pKa = 3.74VEE103 pKa = 4.23ALDD106 pKa = 3.9EE107 pKa = 5.16DD108 pKa = 4.24GDD110 pKa = 3.88ARR112 pKa = 11.84VYY114 pKa = 10.43QGSEE118 pKa = 3.63ADD120 pKa = 3.41EE121 pKa = 4.22TVYY124 pKa = 10.54IDD126 pKa = 4.66PDD128 pKa = 3.83TTSEE132 pKa = 3.87FSILMGSGGADD143 pKa = 2.94TVVTGLGQNISTTDD157 pKa = 3.35GADD160 pKa = 3.34ADD162 pKa = 4.34AEE164 pKa = 4.23EE165 pKa = 5.18ADD167 pKa = 4.32TVRR170 pKa = 11.84LIYY173 pKa = 10.38TPGASEE179 pKa = 4.33EE180 pKa = 4.12LFEE183 pKa = 5.87DD184 pKa = 4.7GSVRR188 pKa = 11.84GSFSMDD194 pKa = 2.97KK195 pKa = 11.06NDD197 pKa = 3.52VLDD200 pKa = 3.6VQFPEE205 pKa = 5.03GEE207 pKa = 4.29SGTLLPVYY215 pKa = 9.42STFVSYY221 pKa = 10.32PRR223 pKa = 11.84DD224 pKa = 3.5ACCSFEE230 pKa = 4.26EE231 pKa = 4.33EE232 pKa = 4.49HH233 pKa = 7.0EE234 pKa = 4.22LTLVYY239 pKa = 10.38IPDD242 pKa = 3.49EE243 pKa = 4.3VEE245 pKa = 3.88YY246 pKa = 10.97SLEE249 pKa = 4.03EE250 pKa = 4.31LEE252 pKa = 4.48SSYY255 pKa = 11.14YY256 pKa = 10.29HH257 pKa = 7.01DD258 pKa = 3.83WEE260 pKa = 4.85ALGMVPIADD269 pKa = 3.69FHH271 pKa = 6.64LGQRR275 pKa = 11.84GEE277 pKa = 3.69IDD279 pKa = 3.46FTHH282 pKa = 6.6EE283 pKa = 4.27GEE285 pKa = 4.26SGEE288 pKa = 3.95FRR290 pKa = 11.84EE291 pKa = 4.02WDD293 pKa = 3.78YY294 pKa = 11.71VRR296 pKa = 11.84EE297 pKa = 4.21PPTVTSNLAIEE308 pKa = 4.12RR309 pKa = 11.84QILIRR314 pKa = 11.84YY315 pKa = 7.21
MM1 pKa = 7.43RR2 pKa = 11.84AAVRR6 pKa = 11.84LQLLPHH12 pKa = 6.62MDD14 pKa = 3.72SIYY17 pKa = 10.54FFCNLTACYY26 pKa = 10.44YY27 pKa = 11.06LLLNRR32 pKa = 11.84GTLVLGTILFAALGVAAIATVISSDD57 pKa = 3.72DD58 pKa = 3.76DD59 pKa = 3.78SHH61 pKa = 8.87EE62 pKa = 4.54EE63 pKa = 3.88IADD66 pKa = 3.66GDD68 pKa = 4.04VPPPIEE74 pKa = 4.83EE75 pKa = 4.08EE76 pKa = 4.32DD77 pKa = 3.83GTPSLIQEE85 pKa = 4.29NGLTVVTGNEE95 pKa = 4.0EE96 pKa = 3.73PVQIDD101 pKa = 3.74VEE103 pKa = 4.23ALDD106 pKa = 3.9EE107 pKa = 5.16DD108 pKa = 4.24GDD110 pKa = 3.88ARR112 pKa = 11.84VYY114 pKa = 10.43QGSEE118 pKa = 3.63ADD120 pKa = 3.41EE121 pKa = 4.22TVYY124 pKa = 10.54IDD126 pKa = 4.66PDD128 pKa = 3.83TTSEE132 pKa = 3.87FSILMGSGGADD143 pKa = 2.94TVVTGLGQNISTTDD157 pKa = 3.35GADD160 pKa = 3.34ADD162 pKa = 4.34AEE164 pKa = 4.23EE165 pKa = 5.18ADD167 pKa = 4.32TVRR170 pKa = 11.84LIYY173 pKa = 10.38TPGASEE179 pKa = 4.33EE180 pKa = 4.12LFEE183 pKa = 5.87DD184 pKa = 4.7GSVRR188 pKa = 11.84GSFSMDD194 pKa = 2.97KK195 pKa = 11.06NDD197 pKa = 3.52VLDD200 pKa = 3.6VQFPEE205 pKa = 5.03GEE207 pKa = 4.29SGTLLPVYY215 pKa = 9.42STFVSYY221 pKa = 10.32PRR223 pKa = 11.84DD224 pKa = 3.5ACCSFEE230 pKa = 4.26EE231 pKa = 4.33EE232 pKa = 4.49HH233 pKa = 7.0EE234 pKa = 4.22LTLVYY239 pKa = 10.38IPDD242 pKa = 3.49EE243 pKa = 4.3VEE245 pKa = 3.88YY246 pKa = 10.97SLEE249 pKa = 4.03EE250 pKa = 4.31LEE252 pKa = 4.48SSYY255 pKa = 11.14YY256 pKa = 10.29HH257 pKa = 7.01DD258 pKa = 3.83WEE260 pKa = 4.85ALGMVPIADD269 pKa = 3.69FHH271 pKa = 6.64LGQRR275 pKa = 11.84GEE277 pKa = 3.69IDD279 pKa = 3.46FTHH282 pKa = 6.6EE283 pKa = 4.27GEE285 pKa = 4.26SGEE288 pKa = 3.95FRR290 pKa = 11.84EE291 pKa = 4.02WDD293 pKa = 3.78YY294 pKa = 11.71VRR296 pKa = 11.84EE297 pKa = 4.21PPTVTSNLAIEE308 pKa = 4.12RR309 pKa = 11.84QILIRR314 pKa = 11.84YY315 pKa = 7.21
Molecular weight: 34.64 kDa
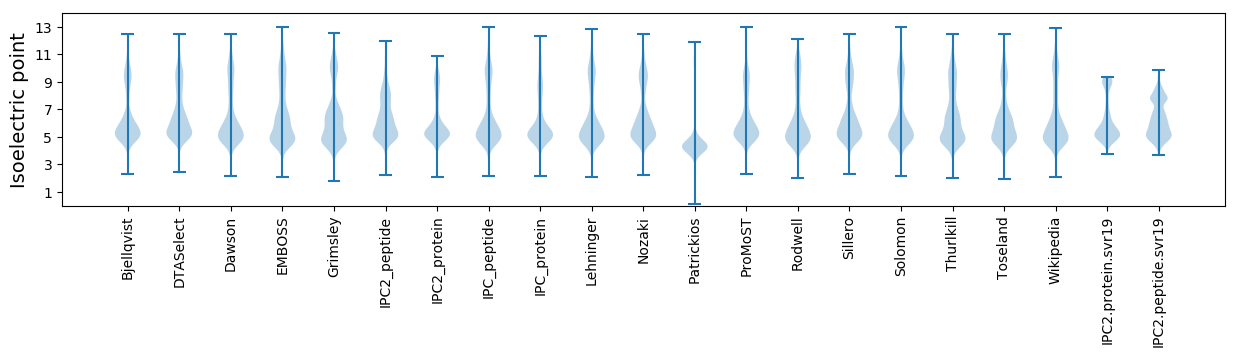
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5LL17|Q5LL17_RUEPO Nitric oxide reductase F protein putative OS=Ruegeria pomeroyi (strain ATCC 700808 / DSM 15171 / DSS-3) OX=246200 GN=SPOA0212 PE=4 SV=1
MM1 pKa = 7.37KK2 pKa = 9.89PVRR5 pKa = 11.84PQPATPPALSEE16 pKa = 4.08NDD18 pKa = 3.41RR19 pKa = 11.84QALNDD24 pKa = 3.81LYY26 pKa = 11.73VRR28 pKa = 11.84GTPEE32 pKa = 3.5NTLRR36 pKa = 11.84AYY38 pKa = 9.92EE39 pKa = 4.3RR40 pKa = 11.84DD41 pKa = 3.82LLYY44 pKa = 10.1LTAWKK49 pKa = 9.83QLSFGAPLAWPEE61 pKa = 3.86QHH63 pKa = 6.25EE64 pKa = 4.39CALRR68 pKa = 11.84FILDD72 pKa = 3.62HH73 pKa = 7.16SRR75 pKa = 11.84DD76 pKa = 3.73LGDD79 pKa = 3.22ATGPARR85 pKa = 11.84EE86 pKa = 4.07VAEE89 pKa = 3.95ALIARR94 pKa = 11.84GLRR97 pKa = 11.84RR98 pKa = 11.84SLACPAPATLNRR110 pKa = 11.84RR111 pKa = 11.84IASWQAFHH119 pKa = 7.42RR120 pKa = 11.84MKK122 pKa = 10.98NLDD125 pKa = 3.79SPFDD129 pKa = 3.9SPLLKK134 pKa = 10.02QARR137 pKa = 11.84AKK139 pKa = 10.41ARR141 pKa = 11.84RR142 pKa = 11.84AAARR146 pKa = 11.84PTAPKK151 pKa = 10.0SRR153 pKa = 11.84NPITRR158 pKa = 11.84PVLEE162 pKa = 4.44QLLATCRR169 pKa = 11.84GSRR172 pKa = 11.84RR173 pKa = 11.84DD174 pKa = 3.53CRR176 pKa = 11.84DD177 pKa = 2.94RR178 pKa = 11.84ALLMLGWASGGRR190 pKa = 11.84RR191 pKa = 11.84RR192 pKa = 11.84SEE194 pKa = 3.65IASLRR199 pKa = 11.84VEE201 pKa = 4.4DD202 pKa = 3.91VSLRR206 pKa = 11.84DD207 pKa = 3.63FAADD211 pKa = 4.81GIAWLMLVEE220 pKa = 4.64TKK222 pKa = 7.05TTRR225 pKa = 11.84RR226 pKa = 11.84GATPRR231 pKa = 11.84LVLKK235 pKa = 10.72GRR237 pKa = 11.84AAQALVHH244 pKa = 5.76WLEE247 pKa = 3.87VSGIEE252 pKa = 4.95DD253 pKa = 3.71GPLFRR258 pKa = 11.84PVSKK262 pKa = 10.35SDD264 pKa = 3.17RR265 pKa = 11.84VLTRR269 pKa = 11.84RR270 pKa = 11.84LTPDD274 pKa = 4.23AIYY277 pKa = 10.34QIVRR281 pKa = 11.84HH282 pKa = 5.82RR283 pKa = 11.84LQLAGLPQDD292 pKa = 3.74FASPHH297 pKa = 5.27GLRR300 pKa = 11.84SGFLTQAALDD310 pKa = 3.96GAPIQAAMRR319 pKa = 11.84LSLHH323 pKa = 6.57RR324 pKa = 11.84SVAQAQRR331 pKa = 11.84YY332 pKa = 8.12YY333 pKa = 11.22DD334 pKa = 3.45DD335 pKa = 4.36VEE337 pKa = 4.36IVEE340 pKa = 4.79NPATDD345 pKa = 3.67LLGG348 pKa = 4.09
MM1 pKa = 7.37KK2 pKa = 9.89PVRR5 pKa = 11.84PQPATPPALSEE16 pKa = 4.08NDD18 pKa = 3.41RR19 pKa = 11.84QALNDD24 pKa = 3.81LYY26 pKa = 11.73VRR28 pKa = 11.84GTPEE32 pKa = 3.5NTLRR36 pKa = 11.84AYY38 pKa = 9.92EE39 pKa = 4.3RR40 pKa = 11.84DD41 pKa = 3.82LLYY44 pKa = 10.1LTAWKK49 pKa = 9.83QLSFGAPLAWPEE61 pKa = 3.86QHH63 pKa = 6.25EE64 pKa = 4.39CALRR68 pKa = 11.84FILDD72 pKa = 3.62HH73 pKa = 7.16SRR75 pKa = 11.84DD76 pKa = 3.73LGDD79 pKa = 3.22ATGPARR85 pKa = 11.84EE86 pKa = 4.07VAEE89 pKa = 3.95ALIARR94 pKa = 11.84GLRR97 pKa = 11.84RR98 pKa = 11.84SLACPAPATLNRR110 pKa = 11.84RR111 pKa = 11.84IASWQAFHH119 pKa = 7.42RR120 pKa = 11.84MKK122 pKa = 10.98NLDD125 pKa = 3.79SPFDD129 pKa = 3.9SPLLKK134 pKa = 10.02QARR137 pKa = 11.84AKK139 pKa = 10.41ARR141 pKa = 11.84RR142 pKa = 11.84AAARR146 pKa = 11.84PTAPKK151 pKa = 10.0SRR153 pKa = 11.84NPITRR158 pKa = 11.84PVLEE162 pKa = 4.44QLLATCRR169 pKa = 11.84GSRR172 pKa = 11.84RR173 pKa = 11.84DD174 pKa = 3.53CRR176 pKa = 11.84DD177 pKa = 2.94RR178 pKa = 11.84ALLMLGWASGGRR190 pKa = 11.84RR191 pKa = 11.84RR192 pKa = 11.84SEE194 pKa = 3.65IASLRR199 pKa = 11.84VEE201 pKa = 4.4DD202 pKa = 3.91VSLRR206 pKa = 11.84DD207 pKa = 3.63FAADD211 pKa = 4.81GIAWLMLVEE220 pKa = 4.64TKK222 pKa = 7.05TTRR225 pKa = 11.84RR226 pKa = 11.84GATPRR231 pKa = 11.84LVLKK235 pKa = 10.72GRR237 pKa = 11.84AAQALVHH244 pKa = 5.76WLEE247 pKa = 3.87VSGIEE252 pKa = 4.95DD253 pKa = 3.71GPLFRR258 pKa = 11.84PVSKK262 pKa = 10.35SDD264 pKa = 3.17RR265 pKa = 11.84VLTRR269 pKa = 11.84RR270 pKa = 11.84LTPDD274 pKa = 4.23AIYY277 pKa = 10.34QIVRR281 pKa = 11.84HH282 pKa = 5.82RR283 pKa = 11.84LQLAGLPQDD292 pKa = 3.74FASPHH297 pKa = 5.27GLRR300 pKa = 11.84SGFLTQAALDD310 pKa = 3.96GAPIQAAMRR319 pKa = 11.84LSLHH323 pKa = 6.57RR324 pKa = 11.84SVAQAQRR331 pKa = 11.84YY332 pKa = 8.12YY333 pKa = 11.22DD334 pKa = 3.45DD335 pKa = 4.36VEE337 pKa = 4.36IVEE340 pKa = 4.79NPATDD345 pKa = 3.67LLGG348 pKa = 4.09
Molecular weight: 38.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1375997 |
30 |
8093 |
322.3 |
34.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.523 ± 0.052 | 0.944 ± 0.012 |
5.919 ± 0.046 | 5.776 ± 0.033 |
3.648 ± 0.025 | 8.94 ± 0.048 |
2.126 ± 0.019 | 4.984 ± 0.024 |
2.849 ± 0.029 | 10.314 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.813 ± 0.023 | 2.522 ± 0.027 |
5.214 ± 0.028 | 3.278 ± 0.021 |
6.971 ± 0.043 | 5.021 ± 0.031 |
5.287 ± 0.032 | 7.193 ± 0.032 |
1.428 ± 0.016 | 2.25 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
